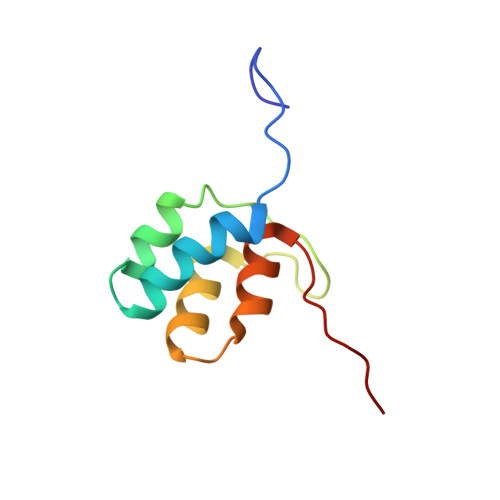
Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase.
Dorival, J., Annaval, T., Risser, F., Collin, S., Roblin, P., Jacob, C., Gruez, A., Chagot, B., Weissman, K.J.(2016) J Am Chem Soc 138: 4155-4167
- PubMed: 26982529
- DOI: https://doi.org/10.1021/jacs.5b13372
- Primary Citation of Related Structures:
2N5D - PubMed Abstract:
Modular polyketide synthases (PKSs) direct the biosynthesis of clinically valuable secondary metabolites in bacteria. The fidelity of chain growth depends on specific recognition between successive subunits in each assembly line: interactions mediated by C- and N-terminal "docking domains" (DDs). We have identified a new family of DDs in trans-acyl transferase PKSs, exemplified by a matched pair from the virginiamycin (Vir) system. In the absence of C-terminal partner (VirA (C)DD) or a downstream catalytic domain, the N-terminal DD (VirFG (N)DD) exhibits multiple characteristics of an intrinsically disordered protein. Fusion of the two docking domains results in a stable fold for VirFG (N)DD and an overall protein-protein complex of unique topology whose structure we support by site-directed mutagenesis. Furthermore, using small-angle X-ray scattering (SAXS), the positions of the flanking acyl carrier protein and ketosynthase domains have been identified, allowing modeling of the complete intersubunit interface.
Organizational Affiliation:
UMR 7365, Ingénierie Moléculaire et Physiopathologie Articulaire (IMoPA), CNRS-Université de Lorraine, Biopôle de l'Université de Lorraine , Campus Biologie Santé, 9 Avenue de la Forêt de Haye, CS 50184, 54505 Vandœuvre-lès-Nancy CEDEX, France.