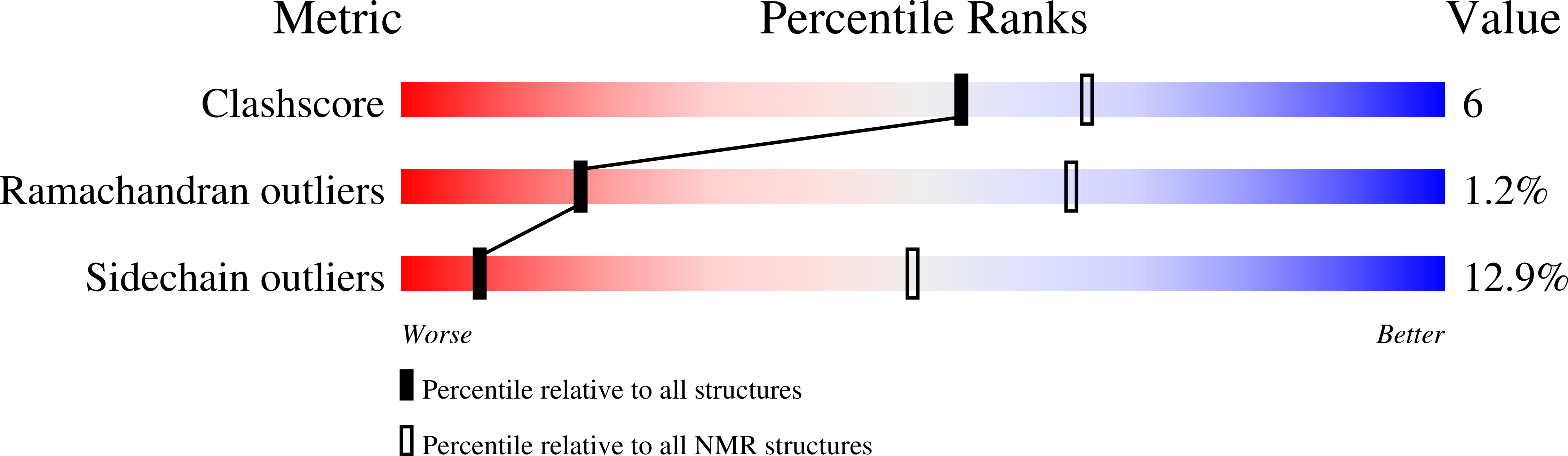Generating Bona Fide Mammalian Prions with Internal Deletions.
Munoz-Montesino, C., Sizun, C., Moudjou, M., Herzog, L., Reine, F., Chapuis, J., Ciric, D., Igel-Egalon, A., Laude, H., Beringue, V., Rezaei, H., Dron, M.(2016) J Virol 90: 6963-6975
- PubMed: 27226369
- DOI: https://doi.org/10.1128/JVI.00555-16
- Primary Citation of Related Structures:
2MV8, 2MV9, 2N53 - PubMed Abstract:
Mammalian prions are PrP proteins with altered structures causing transmissible fatal neurodegenerative diseases. They are self-perpetuating through formation of beta-sheet-rich assemblies that seed conformational change of cellular PrP. Pathological PrP usually forms an insoluble protease-resistant core exhibiting beta-sheet structures but no more alpha-helical content, loosing the three alpha-helices contained in the correctly folded PrP. The lack of a high-resolution prion structure makes it difficult to understand the dynamics of conversion and to identify elements of the protein involved in this process. To determine whether completeness of residues within the protease-resistant domain is required for prions, we performed serial deletions in the helix H2 C terminus of ovine PrP, since this region has previously shown some tolerance to sequence changes without preventing prion replication. Deletions of either four or five residues essentially preserved the overall PrP structure and mutant PrP expressed in RK13 cells were efficiently converted into bona fide prions upon challenge by three different prion strains. Remarkably, deletions in PrP facilitated the replication of two strains that otherwise do not replicate in this cellular context. Prions with internal deletion were self-propagating and de novo infectious for naive homologous and wild-type PrP-expressing cells. Moreover, they caused transmissible spongiform encephalopathies in mice, with similar biochemical signatures and neuropathologies other than the original strains. Prion convertibility and transfer of strain-specific information are thus preserved despite shortening of an alpha-helix in PrP and removal of residues within prions. These findings provide new insights into sequence/structure/infectivity relationship for prions. Prions are misfolded PrP proteins that convert the normal protein into a replicate of their own abnormal form. They are responsible for invariably fatal neurodegenerative disorders. Other aggregation-prone proteins appear to have a prion-like mode of expansion in brains, such as in Alzheimer's or Parkinson's diseases. To date, the resolution of prion structure remains elusive. Thus, to genetically define the landscape of regions critical for prion conversion, we tested the effect of short deletions. We found that, surprisingly, removal of a portion of PrP, the C terminus of alpha-helix H2, did not hamper prion formation but generated infectious agents with an internal deletion that showed characteristics essentially similar to those of original infecting strains. Thus, we demonstrate that completeness of the residues inside prions is not necessary for maintaining infectivity and the main strain-specific information, while reporting one of the few if not the only bona fide prions with an internal deletion.
Organizational Affiliation:
INRA U892, Virologie et Immunologie Moléculaires, Jouy-en-Josas, France.