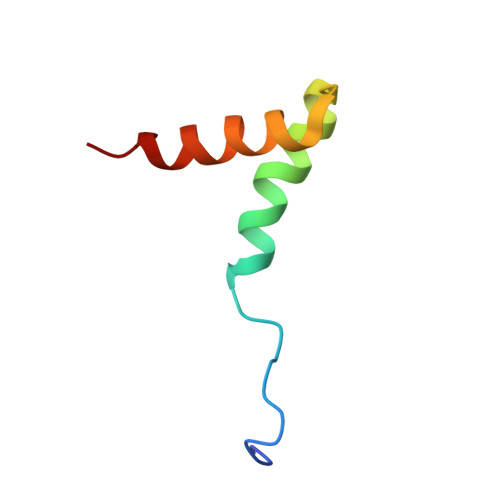
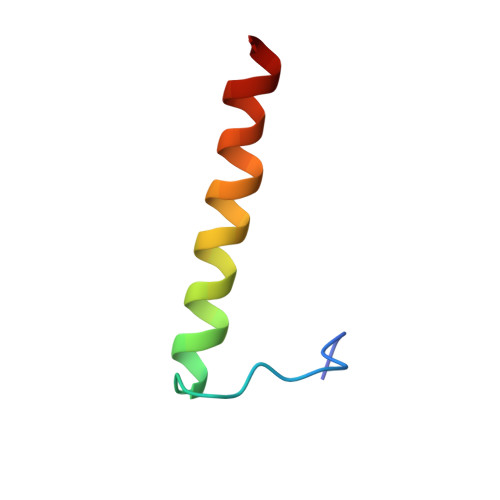
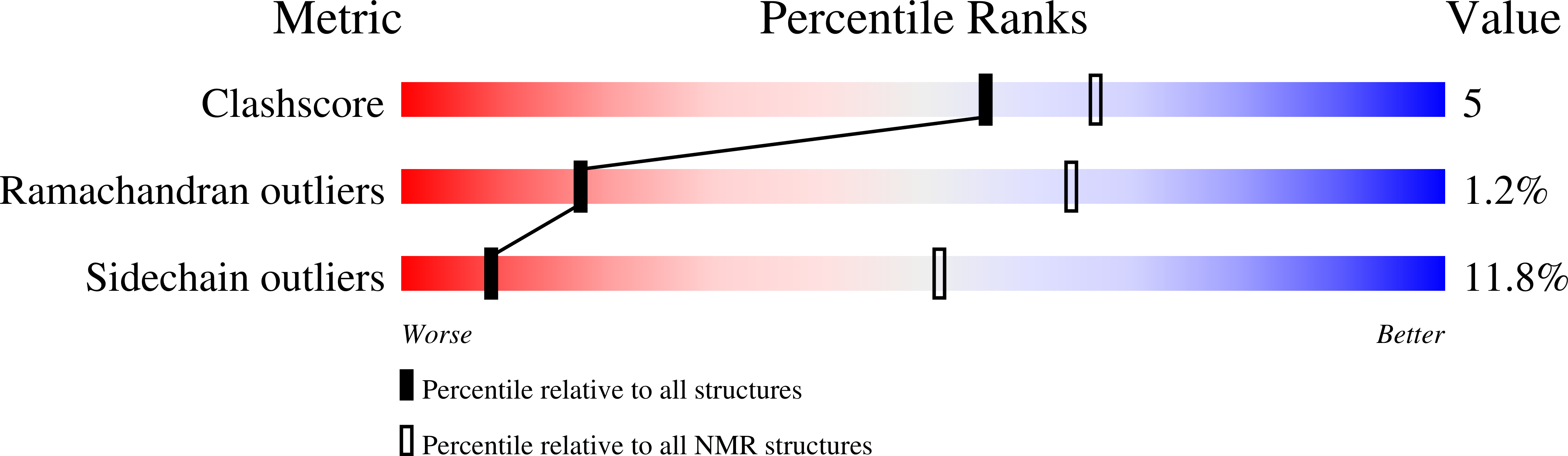Structure of the AML1-ETO NHR3-PKA(RIIalpha) complex and its contribution to AML1-ETO activity.
Corpora, T., Roudaia, L., Oo, Z.M., Chen, W., Manuylova, E., Cai, X., Chen, M.J., Cierpicki, T., Speck, N.A., Bushweller, J.H.(2010) J Mol Biol 402: 560-577
- PubMed: 20708017
- DOI: https://doi.org/10.1016/j.jmb.2010.08.007
- Primary Citation of Related Structures:
2KYG - PubMed Abstract:
AML1-ETO is the chimeric protein product of t(8;21) in acute myeloid leukemia. The ETO portion of the fusion protein includes the nervy homology region (NHR) 3 domain, which shares homology with A-kinase anchoring proteins and interacts with the regulatory subunit of type II cAMP-dependent protein kinase A (PKA(RIIα)). We determined the solution structure of a complex between the AML1-ETO NHR3 domain and PKA(RIIα). Based on this structure, a key residue in AML1-ETO for PKA(RIIα) association was mutated. This mutation did not disrupt AML1-ETO's ability to enhance the clonogenic capacity of primary mouse bone marrow cells or its ability to repress proliferation or granulocyte differentiation. Introduction of the mutation into AML1-ETO had minimal impact on in vivo leukemogenesis. Therefore, the NHR3-PKA(RIIα) protein interaction does not appear to significantly contribute to AML1-ETO's ability to induce leukemia.
Organizational Affiliation:
Department of Chemistry, University of Virginia, Charlottesville, VA 22906, USA.