A Previously Unobserved Conformation for the Human Pex5P Receptor Suggests Roles for Intrinsic Flexibility and Rigid Domain Motions in Ligand Binding
Stanley, W.A., Pursiainen, N., Garman, E.F., Juffer, A., Wilmanns, M., Kursula, P.(2007) BMC Struct Biol 7: 24
- PubMed: 17428317
- DOI: https://doi.org/10.1186/1472-6807-7-24
- Primary Citation of Related Structures:
2J9Q - PubMed Abstract:
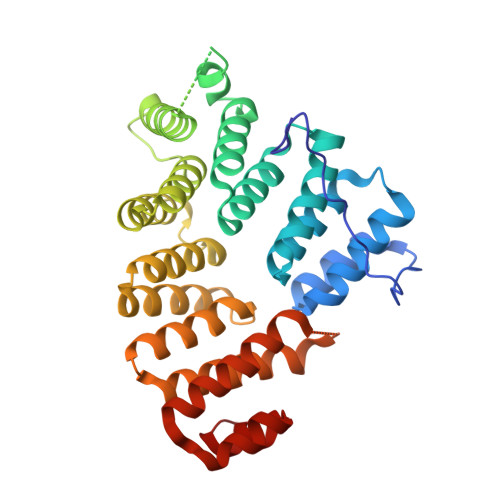
The C-terminal tetratricopeptide (TPR) repeat domain of Pex5p recognises proteins carrying a peroxisomal targeting signal type 1 (PTS1) tripeptide in their C-terminus. Previously, structural data have been obtained from the TPR domain of Pex5p in both the liganded and unliganded states, indicating a conformational change taking place upon cargo protein binding. Such a conformational change would be expected to play a major role both during PTS1 protein recognition as well as in cargo release into the peroxisomal lumen. However, little information is available on the factors that may regulate such structural changes. We have used a range of biophysical and computational methods to further analyse the conformational flexibility and ligand binding of Pex5p. A new crystal form for the human Pex5p C-terminal domain (Pex5p(C)) was obtained in the presence of Sr2+ ions, and the structure presents a novel conformation, distinct from all previous liganded and apo crystal structures for Pex5p(C). The difference relates to a near-rigid body movement of two halves of the molecule, and this movement is different from that required to reach a ring-like conformation upon PTS1 ligand binding. The bound Sr2+ ion changes the dynamic properties of Pex5p(C) affecting its conformation, possibly by making the Sr2+-binding loop - located near the hinge region for the observed domain motions - more rigid. The current data indicate that Pex5p(C) is able to sample a range of conformational states in the absence of bound PTS1 ligand. The domain movements between various apo conformations are distinct from those involved in ligand binding, although the differences between all observed conformations so far can be characterised by the movement of the two halves of Pex5p(C) as near-rigid bodies with respect to each other.
Organizational Affiliation:
EMBL-Hamburg, c/o DESY, Hamburg, Germany. stanley@ccmb.res.in