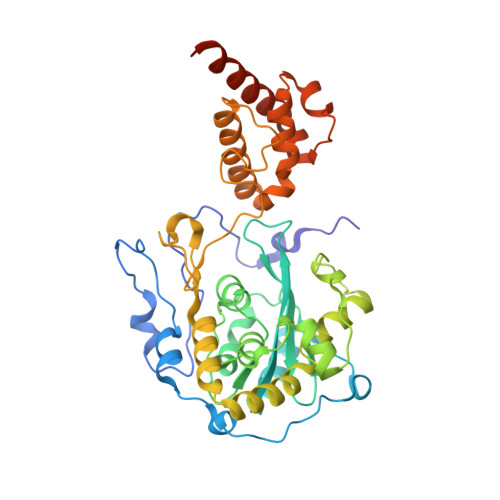
Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Midtgaard, S.F., Assenholt, J., Jonstrup, A.T., Van, L.B., Jensen, T.H., Brodersen, D.E.(2006) Proc Natl Acad Sci U S A 103: 11898-11903
- PubMed: 16882719
- DOI: https://doi.org/10.1073/pnas.0604731103
- Primary Citation of Related Structures:
2HBJ, 2HBK, 2HBL, 2HBM - PubMed Abstract:
The multisubunit eukaryotic exosome is an essential RNA processing and degradation machine. In its nuclear form, the exosome associates with the auxiliary factor Rrp6p, which participates in both RNA processing and degradation reactions. The crystal structure of Saccharomyces cerevisiae Rrp6p displays a conserved RNase D core with a flanking HRDC (helicase and RNase D C-terminal) domain in an unusual conformation shown to be important for the processing function of the enzyme. Complexes with AMP and UMP, the products of the RNA degradation process, reveal how the protein specifically recognizes ribonucleotides and their bases. Finally, in vivo mutational studies show the importance of the domain contacts for the processing function of Rrp6p and highlight fundamental differences between the protein and its prokaryotic RNase D counterparts.
Organizational Affiliation:
Centre for Structural Biology, Department of Molecular Biology, University of Aarhus, Gustav Wieds Vej 10c, DK-8000 Aarhus C, Denmark.