An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
Yin, J., Cherney, M.M., Bergmann, E.M., Zhang, J., Huitema, C., Pettersson, H., Eltis, L.D., Vederas, J.C., James, M.N.(2006) J Mol Biol 361: 673-686
- PubMed: 16860823
- DOI: https://doi.org/10.1016/j.jmb.2006.06.047
- Primary Citation of Related Structures:
2H6M, 2H9H, 2HAL - PubMed Abstract:
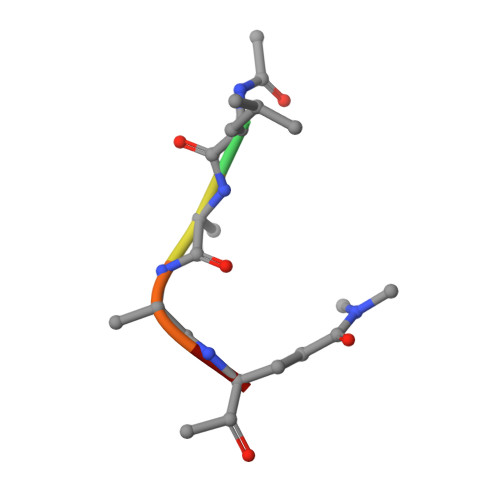
We have solved the crystal and molecular structures of hepatitis A viral (HAV) 3C proteinase, a cysteine peptidase having a chymotrypsin-like protein fold, in complex with each of three tetrapeptidyl-based methyl ketone inhibitors to resolutions beyond 1.4 A, the highest resolution to date for a 3C or a 3C-Like (e.g. SARS viral main proteinase) peptidase. The residues of the beta-hairpin motif (residues 138-158), an extension of two beta-strands of the C-terminal beta-barrel of HAV 3C are critical for the interactions between the enzyme and the tetrapeptide portion of these inhibitors that are analogous to the residues at the P4 to P1 positions in the natural substrates of picornaviral 3C proteinases. Unexpectedly, the Sgamma of Cys172 forms two covalent bonds with each inhibitor, yielding an unusual episulfide cation (thiiranium ring) stabilized by a nearby oxyanion. This result suggests a mechanism of inactivation of 3C peptidases by methyl ketone inhibitors that is distinct from that occurring in the structurally related serine proteinases or in the papain-like cysteine peptidases. It also provides insight into the mechanisms underlying both the inactivation of HAV 3C by these inhibitors and on the proteolysis of natural substrates by this viral cysteine peptidase.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, AB, Canada T6G 2H7.