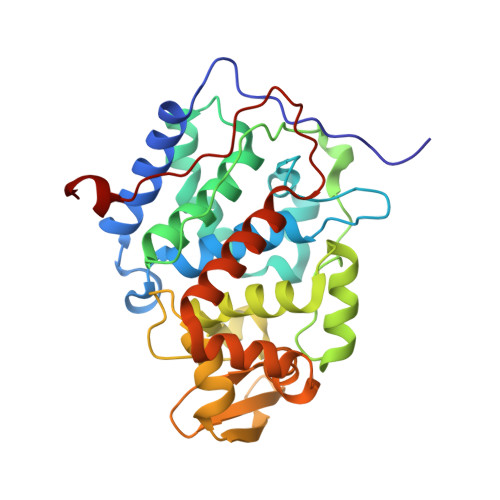
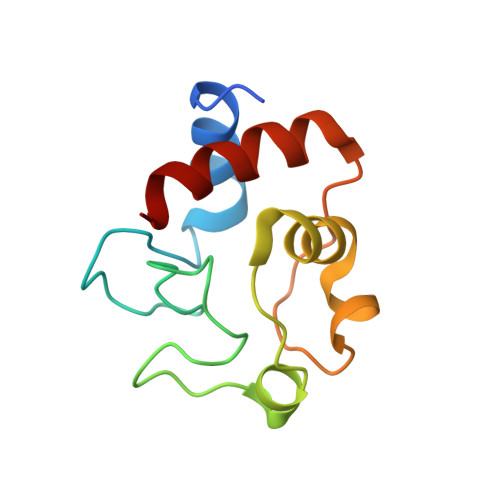
Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR.
Volkov, A.N., Worrall, J.A., Holtzmann, E., Ubbink, M.(2006) Proc Natl Acad Sci U S A 103: 18945-18950
- PubMed: 17146057
- DOI: https://doi.org/10.1073/pnas.0603551103
- Primary Citation of Related Structures:
2GB8 - PubMed Abstract:
The physiological complex of yeast cytochrome c peroxidase and iso-1-cytochrome c is a paradigm for biological electron transfer. Using paramagnetic NMR spectroscopy, we have determined the conformation of the protein complex in solution, which is shown to be very similar to that observed in the crystal structure [Pelletier H, Kraut J (1992) Science 258:1748-1755]. Our results support the view that this transient electron transfer complex is dynamic. The solution structure represents the dominant protein-protein orientation, which, according to our estimates, is occupied for >70% of the lifetime of the complex, with the rest of the time spent in the dynamic encounter state. Based on the observed paramagnetic effects, we have delineated the conformational space sampled by the protein molecules during the dynamic part of the interaction, providing experimental support for the theoretical predictions of the classical Brownian dynamics study [Northrup SH, Boles JO, Reynolds JCL (1988) Science 241:67-70]. Our findings corroborate the dynamic behavior of this complex and offer an insight into the mechanism of the protein complex formation in solution.
Organizational Affiliation:
Leiden Institute of Chemistry, Leiden University, Gorlaeus Laboratories, P.O. Box 9502, 2300 RA Leiden, The Netherlands.