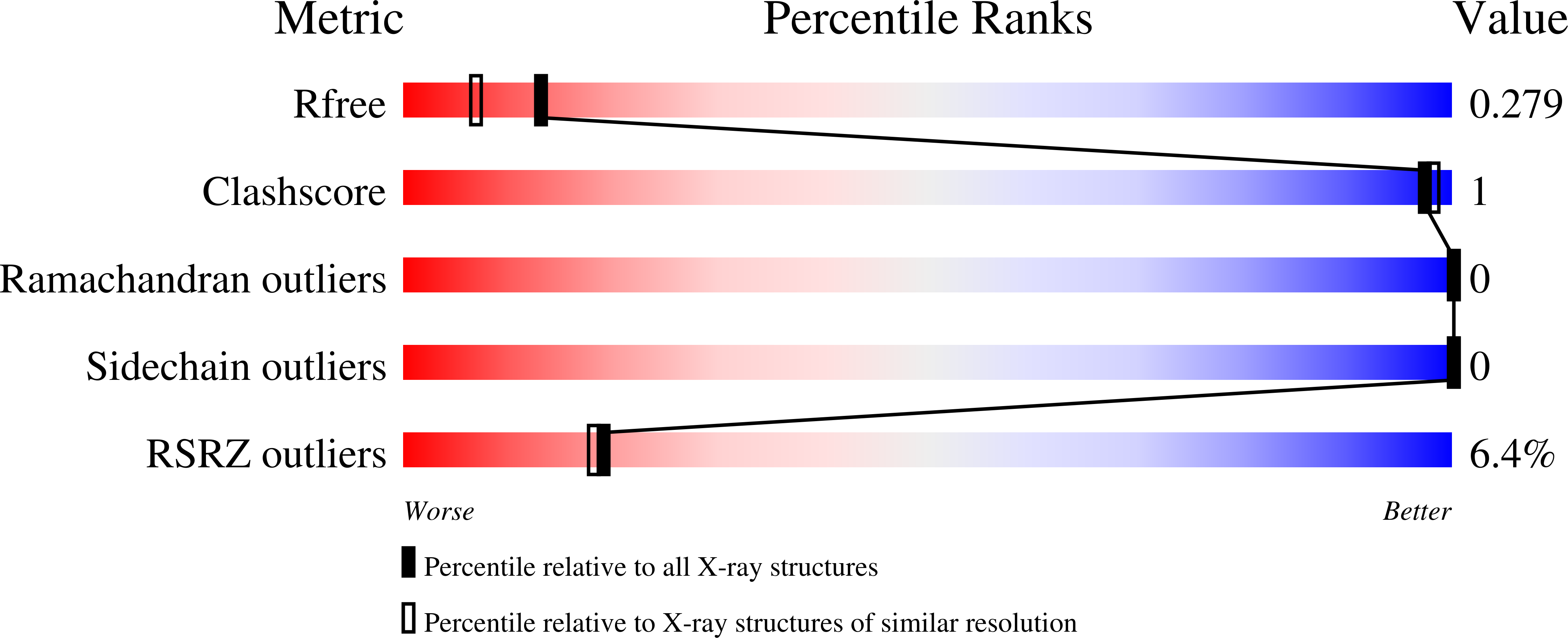
Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition
Mishima, M., Maesaki, R., Kasa, M., Watanabe, T., Fukata, M., Kaibuchi, K., Hakoshima, T.(2007) Proc Natl Acad Sci U S A 104: 10346-10351
- PubMed: 17563362
- DOI: https://doi.org/10.1073/pnas.0703876104
- Primary Citation of Related Structures:
2E3H, 2E3I, 2E4H - PubMed Abstract:
Cytoplasmic linker protein 170 (CLIP-170) is a prototype of the plus end-tracking proteins that regulate microtubule dynamics, but it is obscure how CLIP-170 recognizes the microtubule plus end and contributes to polymerization rescue. Crystallographic, NMR, and mutation studies of two tandem cytoskeleton-associated protein glycine-rich (CAP-Gly) domains of CLIP-170, CAP-Gly-1 and CAP-Gly-2, revealed positively charged basic grooves of both CAP-Gly domains for tubulin binding, whereas the CAP-Gly-2 domain possesses a more basic groove and directly binds the EExEEY/F motif of the C-terminal acidic-tail ends of alpha-tubulin. Notably, the p150(Glued) CAP-Gly domain that is furnished with a less positively charged surface only weakly interacts with the alpha-tubulin acidic tail. Mutation studies showed that this acidic sextette motif is the minimum region for CAP-Gly binding. The C-terminal zinc knuckle domains of CLIP-170 bind the basic groove to inhibit the binding to the acidic tails. These results provide a structural basis for the proposed CLIP-170 copolymerization with tubulin on the microtubule plus end. CLIP-170 strongly binds the acidic tails of EB1 as well as those of alpha-tubulins, indicating that EB1 localized at the plus end contributes to CLIP-170 recruitment to the plus end. We suggest that CLIP-170 stimulates microtubule polymerization and/or nucleation by neutralizing the negative charges of tubulins with the highly positive charges of the CLIP-170 CAP-Gly domains. Once CLIP-170 binds microtubule, the released zinc knuckle domain may serve to recruit dynein to the plus end by interacting with p150(Glued) and LIS1. Thus, our structures provide the structural basis for the specific dynein loading on the microtubule plus end.
Organizational Affiliation:
Graduate School of Biological Science, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.