The refined crystal structure of alpha-cobratoxin from Naja naja siamensis at 2.4-A resolution.
Betzel, C., Lange, G., Pal, G.P., Wilson, K.S., Maelicke, A., Saenger, W.(1991) J Biol Chem 266: 21530-21536
- PubMed: 1939183
- DOI: https://doi.org/10.2210/pdb2ctx/pdb
- Primary Citation of Related Structures:
2CTX - PubMed Abstract:
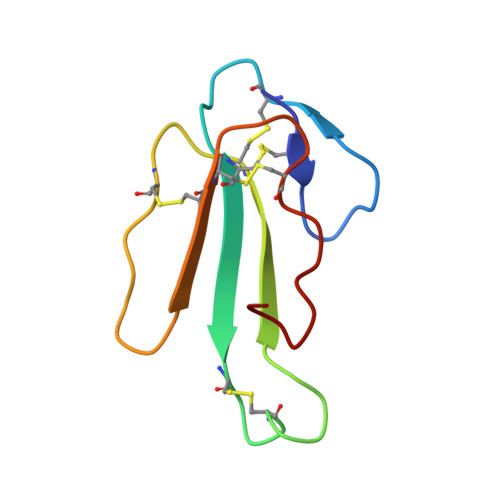
The crystal structure of the "long" alpha-neurotoxin alpha-cobratoxin was refined to an R-factor of 19.5% using 3271 x-ray data to 2.4-A resolution. The polypeptide chain forms three loops, I, II, III, knotted together by four disulfide bridges, with the most prominent, loop II, containing another disulfide close to its lower tip. Loop I is stabilized by one beta-turn and two beta-sheet hydrogen bonds; loop II by eight beta-sheet hydrogen bonds, with the tip folded into two distorted right-handed helical turns stabilized by two alpha-helical and two beta-turn hydrogen bonds; and loop III by hydrophobic interactions and one beta-turn. Loop II and one strand of loop III form an antiparallel triple-pleated beta-sheet, and tight anchoring of the Asn63 side chain fixes the tail segment. In the crystal lattice, the alpha-cobratoxin molecules dimerize by beta-sheet formation between strands 53 and 57 of symmetry-related molecules. Because such interactions are found also in a cardiotoxin and alpha-bungarotoxin, this could be of importance for interaction with acetylcholine receptor.
Organizational Affiliation:
European Molecular Biology Laboratory, Deutsches Elektronen Synchrotron, Hamburg, Federal Republic of Germany.