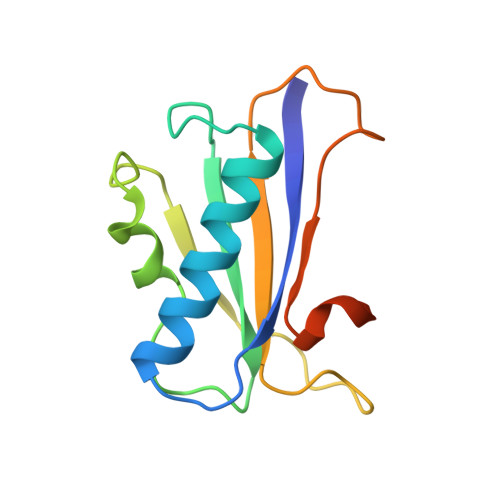
Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog.
Chook, Y.M., Ke, H., Lipscomb, W.N.(1993) Proc Natl Acad Sci U S A 90: 8600-8603
- PubMed: 8378335
- DOI: https://doi.org/10.1073/pnas.90.18.8600
- Primary Citation of Related Structures:
2CHS, 2CHT - PubMed Abstract:
We have solved the structure of a chorismate mutase (chorismate pyruvatemutase, EC 5.4.99.5), the 1.9-A crystal structure of the monofunctional enzyme from Bacillus subtilis. The structure determination process was an unusual one, involving 12 monomers of the enzyme in the asymmetric unit. This structure was solved by the multiple isomorphous replacement method with partial structure phase combination and molecular averaging. The final model, which includes 1380 residues and 522 water molecules in an asymmetric unit, has been refined at 1.9 A and the current crystallographic R value is 0.201. The B. subtilis chorismate mutase is a homotrimer, with beta-sheets from each monomer packing to form the core of a pseudo-alpha beta-barrel with helices on the outside of the trimer. In addition, the active sites have been located by using data from a complex with an endo-oxabicyclic inhibitor that mimics the transition state of the reaction. The structure of this complex has been refined to 2.2 A with a current R value of 0.182 for a model that includes 1388 residues, 12 inhibitor molecules, and 530 water molecules in the asymmetric unit. In each trimer, three equivalent active sites are located at the interfaces of two adjacent subunits.
Organizational Affiliation:
Gibbs Chemical Laboratory, Harvard University, Cambridge, MA 02138.