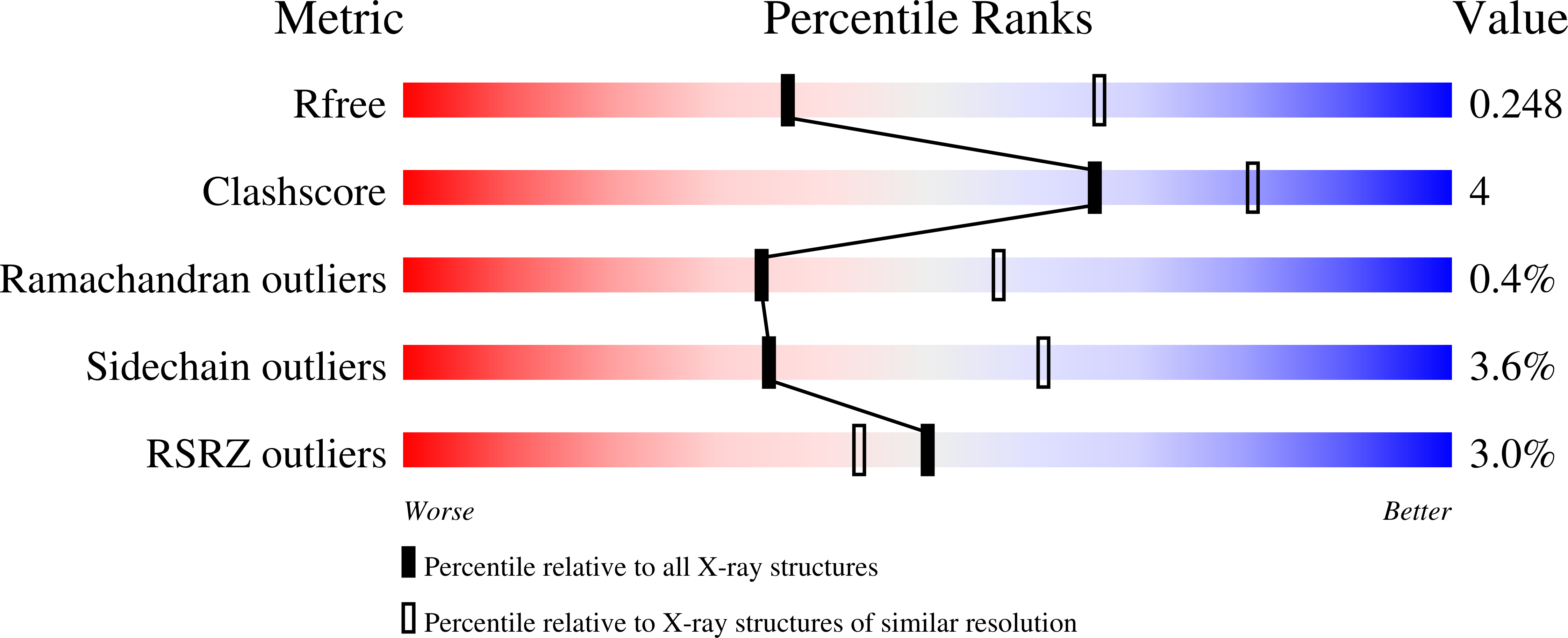
Allosteric Threonine Synthase: Reorganization of the Pyridoxal Phosphate Site Upon Asymmetric Activation Through S-Adenosylmethionine Binding to a Novel Site.
Mas-Droux, C., Biou, V., Dumas, R.(2006) J Biol Chem 281: 5188
- PubMed: 16319072
- DOI: https://doi.org/10.1074/jbc.M509798200
- Primary Citation of Related Structures:
2C2B, 2C2G - PubMed Abstract:
Threonine synthase (TS) is a fold-type II pyridoxal phosphate (PLP)-dependent enzyme that catalyzes the ultimate step of threonine synthesis in plants and microorganisms. Unlike the enzyme from microorganisms, plant TS is activated by S-adenosylmethionine (AdoMet). The mechanism of activation has remained unknown up to now. We report here the crystallographic structures of Arabidopsis thaliana TS in complex with PLP (aTS) and with PLP and AdoMet (aTS-AdoMet), which show with atomic detail how AdoMet activates TS. The aTS structure reveals a PLP orientation never previously observed for a type II PLP-dependent enzyme and explains the low activity of plant TS in the absence of its allosteric activator. The aTS-AdoMet structure shows that activation of the enzyme upon AdoMet binding triggers a large reorganization of active site loops in one monomer of the structural dimer and allows the displacement of PLP to its active conformation. Comparison with other TS structures shows that activation of the second monomer may be triggered by substrate binding. This structure also discloses a novel fold for two AdoMet binding sites located at the dimer interface, each site containing two AdoMet effectors bound in tandem. Moreover, aTS-AdoMet is the first structure of an enzyme that uses AdoMet as an allosteric effector.
Organizational Affiliation:
Laboratoire de Physiologie Cellulaire Végétale, Département Réponse et Dynamique Cellulaires, CNRS Commissariat à l'Energie Atomique, 38054 Grenoble, France.