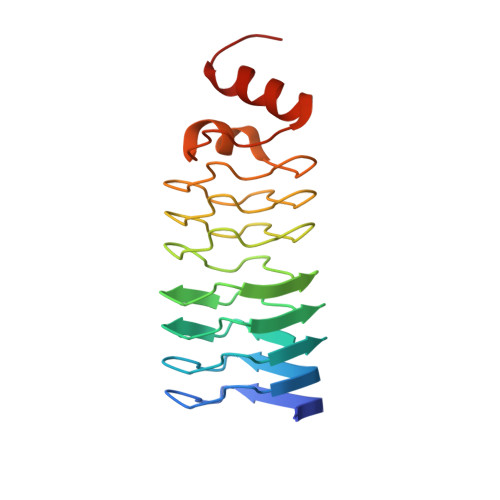
A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Hegde, S.S., Vetting, M.W., Roderick, S.L., Mitchenall, L.A., Maxwell, A., Takiff, H.E., Blanchard, J.S.(2005) Science 308: 1480
- PubMed: 15933203
- DOI: https://doi.org/10.1126/science.1110699
- Primary Citation of Related Structures:
2BM4, 2BM5, 2BM6, 2BM7 - PubMed Abstract:
Fluoroquinolones are gaining increasing importance in the treatment of tuberculosis. The expression of MfpA, a member of the pentapeptide repeat family of proteins from Mycobacterium tuberculosis, causes resistance to ciprofloxacin and sparfloxacin. This protein binds to DNA gyrase and inhibits its activity. Its three-dimensional structure reveals a fold, which we have named the right-handed quadrilateral beta helix, that exhibits size, shape, and electrostatic similarity to B-form DNA. This represents a form of DNA mimicry and explains both its inhibitory effect on DNA gyrase and fluoroquinolone resistance resulting from the protein's expression in vivo.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA.