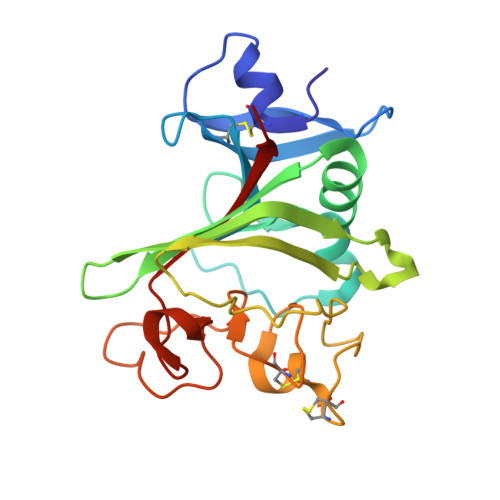
Structure of the angiopoietin-2 receptor binding domain and identification of surfaces involved in Tie2 recognition.
Barton, W.A., Tzvetkova, D., Nikolov, D.B.(2005) Structure 13: 825-832
- PubMed: 15893672
- DOI: https://doi.org/10.1016/j.str.2005.03.009
- Primary Citation of Related Structures:
1Z3S, 1Z3U - PubMed Abstract:
The angiopoietins comprise a small class of secreted glycoproteins that play crucial roles in the maturation and maintenance of the mammalian vascular and lymphatic systems. They exert their effects through a member of the tyrosine kinase receptor family, Tie2. Angiopoietin/Tie2 signaling is unique among tyrosine kinase receptor-ligand systems in that distinct angiopoietin ligands, although highly homologous, can function as agonists or antagonists in a context-dependent manner. In an effort to understand this molecular dichotomy, we have crystallized and determined the 2.4 A crystal structure of the Angiopoietin-2 (Ang2) receptor binding region. The structure reveals a fibrinogen fold with a unique C-terminal P domain. Conservation analysis and structure-based mutagenesis identify a groove on the Ang2 molecular surface that mediates receptor recognition.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, New York 10021, USA.