Isolation, gene expression and solution structure of a novel moricin analogue, antibacterial peptide from a lepidopteran insect, Spodoptera litura
Oizumi, Y., Hemmi, H., Minami, M., Asaoka, A., Yamakawa, M.(2005) Biochim Biophys Acta 1752: 83-92
- PubMed: 16115804
- DOI: https://doi.org/10.1016/j.bbapap.2005.07.013
- Primary Citation of Related Structures:
1X22 - PubMed Abstract:
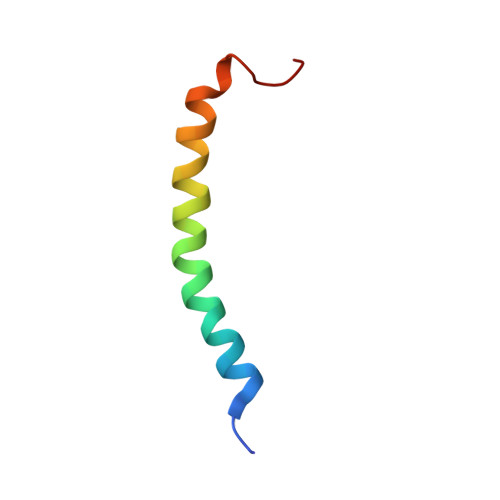
An antibacterial peptide was isolated from a lepidopteran insect, Spodoptera litura. The molecular mass of this peptide was determined to be 4489.55 by matrix assisted laser desorption/ionization-time of flight mass (MALDI-TOF MS) spectrometry. The peptide consists of 42 amino acids and the sequence has 69-98% identity to those of moricin-related peptides, antibacterial peptides from lepidopetran insects. Thus, the peptide was designated S. litura (Sl) moricin. Sl moricin showed a broad antibacterial spectrum against Gram-positive and negative bacteria. Sl moricin gene was inducible by bacterial injection and expressed tissue-specifically in the fat body and hemocytes. Furthermore, the solution structure of Sl moricin was determined by two-dimensional (2D) 1H-nuclear magnetic resonance (NMR) spectroscopy and hybrid distance geometry-simulated annealing calculation. The tertiary structure revealed a long alpha-helix containing eight turns along nearly the full length of the peptide like that of moricin, confirming that Sl moricin is a new moricin-like antibacterial peptide. These results suggest that moricin is present not only in B. mori but also in other lepidopteran insects forming a gene family.
Organizational Affiliation:
Biotechnology Research Center, Kubota Corporation, Ryugasaki, Ibaraki 301-0852, Japan. scott.roberts@bbsrc.ac.uk