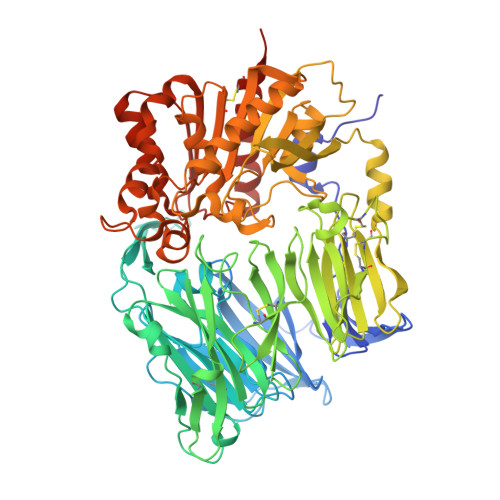
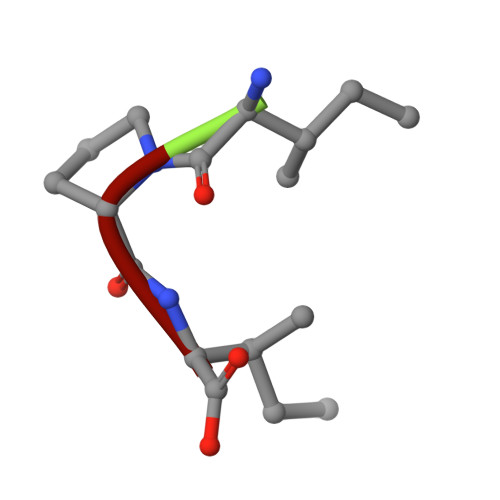
The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A
Hiramatsu, H., Yamamoto, A., Kyono, K., Higashiyama, Y., Fukushima, C., Shima, H., Sugiyama, S., Inaka, K., Shimizu, R.(2004) Biol Chem 385: 561-564
- PubMed: 15255191
- DOI: https://doi.org/10.1515/BC.2004.068
- Primary Citation of Related Structures:
1WCY - PubMed Abstract:
Dipeptidyl peptidase IV (DPPIV) is a serine protease, a member of the prolyl oligopeptidase (POP) family, and has been implicated in several diseases. Therefore, it seems important to develop selective inhibitors for human DPPIV (hDPPIV) that are able to control the biological function of hDPPIV. In order to elucidate the binding mode and substrate specificity, we determined the crystal structure complex of hDPPIV and diprotin A (IIe-Pro-IIe), a slowly hydrolyzed substrate of hDPPIV, at 2.2 A resolution. In this paper, we discuss the molecular interaction mechanism of diprotin A with hDPPIV based on the X-ray crystal structure.
Organizational Affiliation:
Tanabe Seiyaku Co. Ltd., 3-16-89 Kashima, Yodogawa-ku, Osaka 532-8505, Japan.