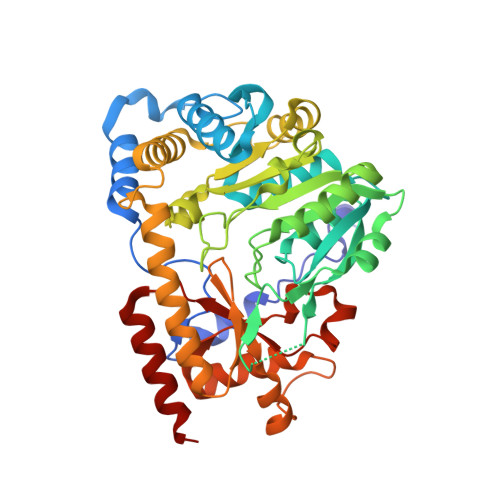
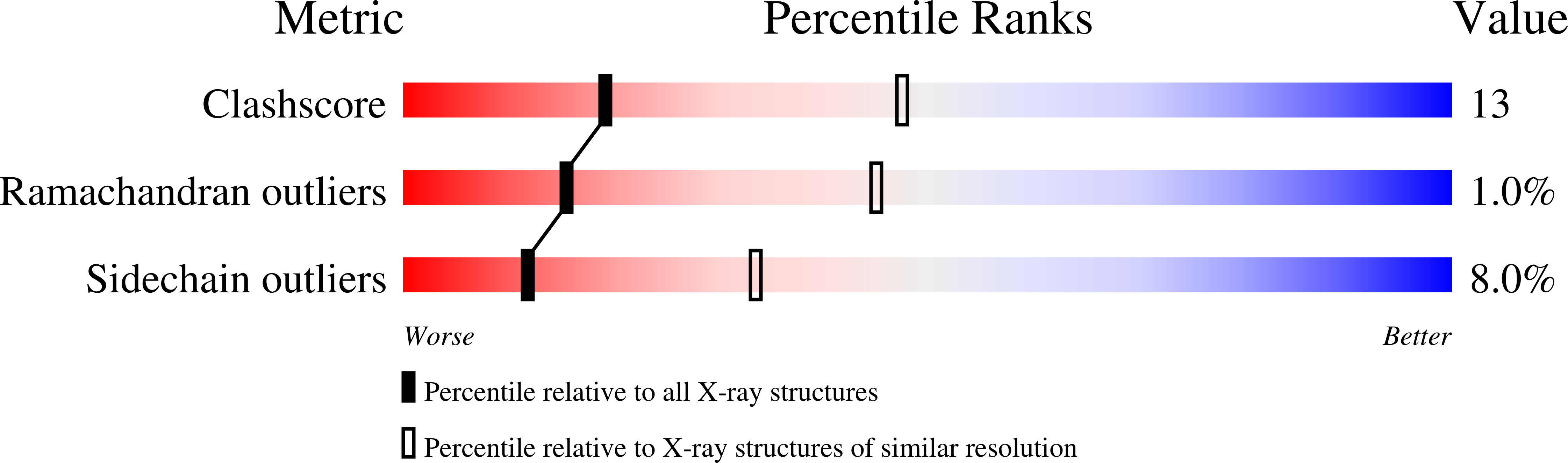Crystal Structure of Human Kynurenine Aminotransferase I
Rossi, F., Han, Q., Li, J., Li, J., Rizzi, M.(2004) J Biol Chem 279: 50214
- PubMed: 15364907
- DOI: https://doi.org/10.1074/jbc.M409291200
- Primary Citation of Related Structures:
1W7L, 1W7M, 1W7N - PubMed Abstract:
The kynurenine pathway has long been regarded as a valuable target for the treatment of several neurological disorders accompanied by unbalanced levels of metabolites along the catabolic cascade, kynurenic acid among them. The irreversible transamination of kynurenine is the sole source of kynurenic acid, and it is catalyzed by different isoforms of the 5'-pyridoxal phosphate-dependent kynurenine aminotransferase (KAT). The KAT-I isozyme has also been reported to possess beta-lyase activity toward several sulfur- and selenium-conjugated molecules, leading to the proposal of a role of the enzyme in carcinogenesis associated with environmental pollutants. We solved the structure of human KAT-I in its 5'-pyridoxal phosphate and pyridoxamine phosphate forms and in complex with the competing substrate l-Phe. The enzyme active site revealed a striking crown of aromatic residues decorating the ligand binding pocket, which we propose as a major molecular determinant for substrate recognition. Ligand-induced conformational changes affecting Tyr(101) and the Trp(18)-bearing alpha-helix H1 appear to play a central role in catalysis. Our data reveal a key structural role of Glu(27), providing a molecular basis for the reported loss of enzymatic activity displayed by the equivalent Glu --> Gly mutation in KAT-I of spontaneously hypertensive rats.
Organizational Affiliation:
Dipartimento di Scienze Chimiche, Alimentari, Farmaceutiche, Farmacologiche-Istituto Nazionale di Fisica della Materia, University of Piemonte Orientale "Amedeo Avogadro," Via Bovio 6, 28100 Novara, Italy.