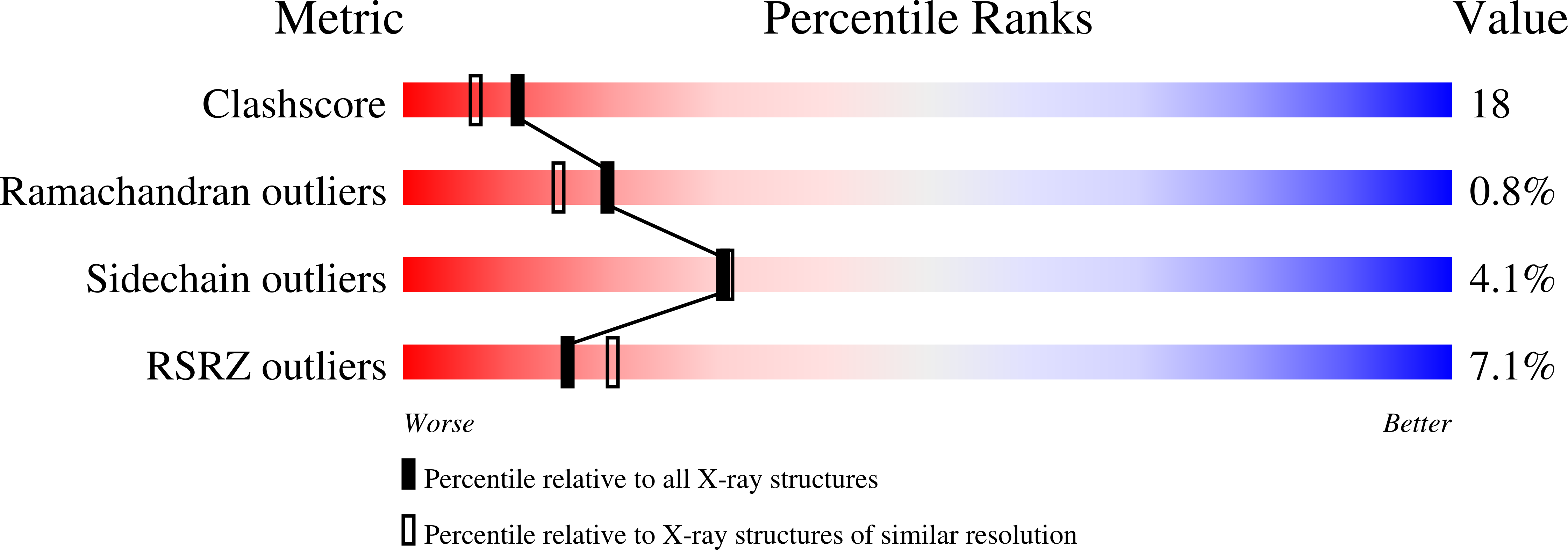Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding
Lopez-Lucendo, M.I.F., Solis, D., Andre, S., Hirabayashi, J., Kasai, K., Kaltner, H., Gabius, H.J., Romero, A.(2004) J Mol Biol 343: 957
- PubMed: 15476813
- DOI: https://doi.org/10.1016/j.jmb.2004.08.078
- Primary Citation of Related Structures:
1GZW, 1W6M, 1W6N, 1W6O, 1W6P, 1W6Q - PubMed Abstract:
Human galectin-1 is a potent multifunctional effector that participates in specific protein-carbohydrate and protein-protein (lipid) interactions. By determining its X-ray structure, we provide the basis to define the structure of its ligand-binding pocket and to perform rational drug design. We have also analysed whether single-site mutations introduced at some distance from the carbohydrate recognition domain can affect the lectin fold and influence sugar binding. Both the substitutions introduced in the C2S and R111H mutants altered the presentation of the loop, harbouring Asp123 in the common "jelly-roll" fold. The orientation of the side-chain was inverted 180 degrees and the positions of two key residues in the sugar-binding site of the R111H mutant were notably shifted, i.e. His52 and Trp68. Titration calorimetry was used to define the decrease in ligand affinity in both mutants and a significant increase in the entropic penalty was found to outweigh a slight enhancement of the enthalpic contribution. The position of the SH-groups in the galectin appeared to considerably restrict the potential to form intramolecular disulphide bridges and was assumed to be the reason for the unstable lectin activity in the absence of reducing agent. However, this offers no obvious explanation for the improved stability of the C2S mutant under oxidative conditions. The noted long-range effects in single-site mutants are relevant for the functional divergence of closely related galectins and in more general terms, the functionality definition of distinct amino acids.
Organizational Affiliation:
Centro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, E-28040 Madrid, Spain.