CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM
Mishra, V., Bilgrami, S., Paramasivam, M., Yadav, S., Sharma, R.S., Kaur, P., Srinivasan, A., Babu, C.R., Singh, T.P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Himalayan mistletoe ribosome-inactivating protein | 240 | Viscum album | Mutation(s): 0 | 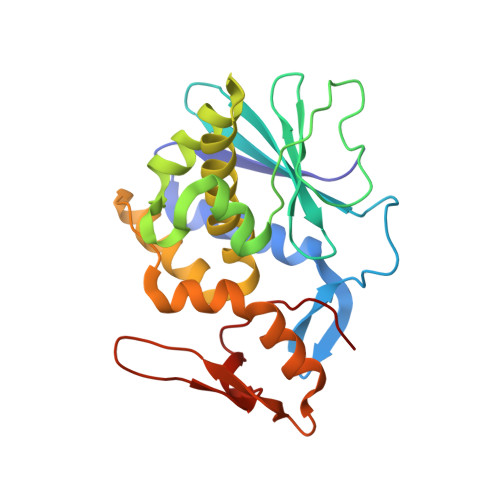 | |
UniProt | |||||
Find proteins for Q6ITZ3 (Viscum album) Explore Q6ITZ3 Go to UniProtKB: Q6ITZ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6ITZ3 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Himalayan mistletoe ribosome-inactivating protein | 255 | Viscum album | Mutation(s): 0 | 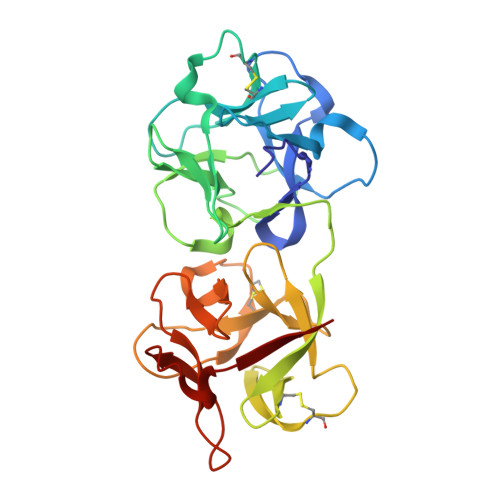 | |
UniProt | |||||
Find proteins for Q6ITZ3 (Viscum album) Explore Q6ITZ3 Go to UniProtKB: Q6ITZ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6ITZ3 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | D | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G31886NL GlyCosmos: G31886NL GlyGen: G31886NL | |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| P6C Query on P6C | H [auth A] | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID C10 H11 N5 O2 GPHPGFBRJYCUDO-UHFFFAOYSA-N |  | ||
| NAG Query on NAG | G [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| GAL Query on GAL | I [auth B] | beta-D-galactopyranose C6 H12 O6 WQZGKKKJIJFFOK-FPRJBGLDSA-N |  | ||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900004 Query on PRD_900004 | F | beta-lactose | Oligosaccharide / Nutrient |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 109.422 | α = 90 |
| b = 109.422 | β = 90 |
| c = 309.798 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |