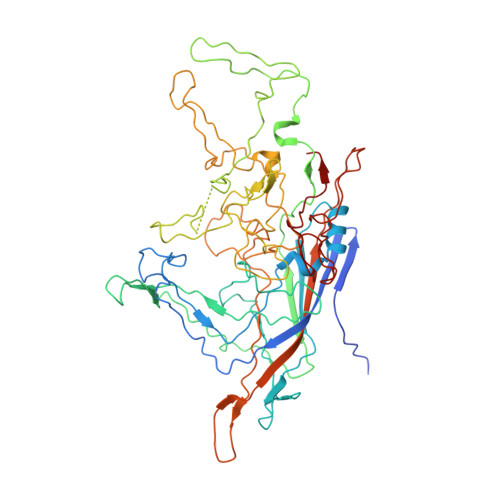
The structure of human parvovirus B19.
Kaufmann, B., Simpson, A.A., Rossmann, M.G.(2004) Proc Natl Acad Sci U S A 101: 11628-11633
- PubMed: 15289612
- DOI: https://doi.org/10.1073/pnas.0402992101
- Primary Citation of Related Structures:
1S58 - PubMed Abstract:
Human parvovirus B19 is the only parvovirus known to be a human pathogen. The structure of recombinant B19-like particles has been determined to approximately 3.5-A resolution by x-ray crystallography and, to our knowledge, represents the first near-atomic structure of an Erythrovirus. The polypeptide fold of the major capsid protein VP2 is a "jelly roll" with a beta-barrel motif similar to that found in many icosahedral viruses. The large loops connecting the strands of the beta-barrel form surface features that differentiate B19 from other parvoviruses. Although B19 VP2 has only 26% sequence identity to VP3 of adeno-associated virus, 72% of the C(alpha) atoms can be aligned structurally with a rms deviation of 1.8 A. Both viruses require an integrin as a coreceptor, and conserved surface features suggest a common receptor-binding region.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, 915 West State Street, West Lafayette, IN 47907-2054, USA.