Structure/Function Studies on a S-Adenosyl-l-methionine-dependent Uroporphyrinogen III C Methyltransferase (SUMT), a Key Regulatory Enzyme of Tetrapyrrole Biosynthesis
Vevodova, J., Graham, R.M., Raux, E., Schubert, H.L., Roper, D.I., Brindley, A.A., Scott, A.I., Roessner, C.A., Stamford, N.P.J., Stroupe, M.E., Getzoff, E.D., Warren, M.J., Wilson, K.S.(2004) J Mol Biol 344: 419-433
- PubMed: 15522295
- DOI: https://doi.org/10.1016/j.jmb.2004.09.020
- Primary Citation of Related Structures:
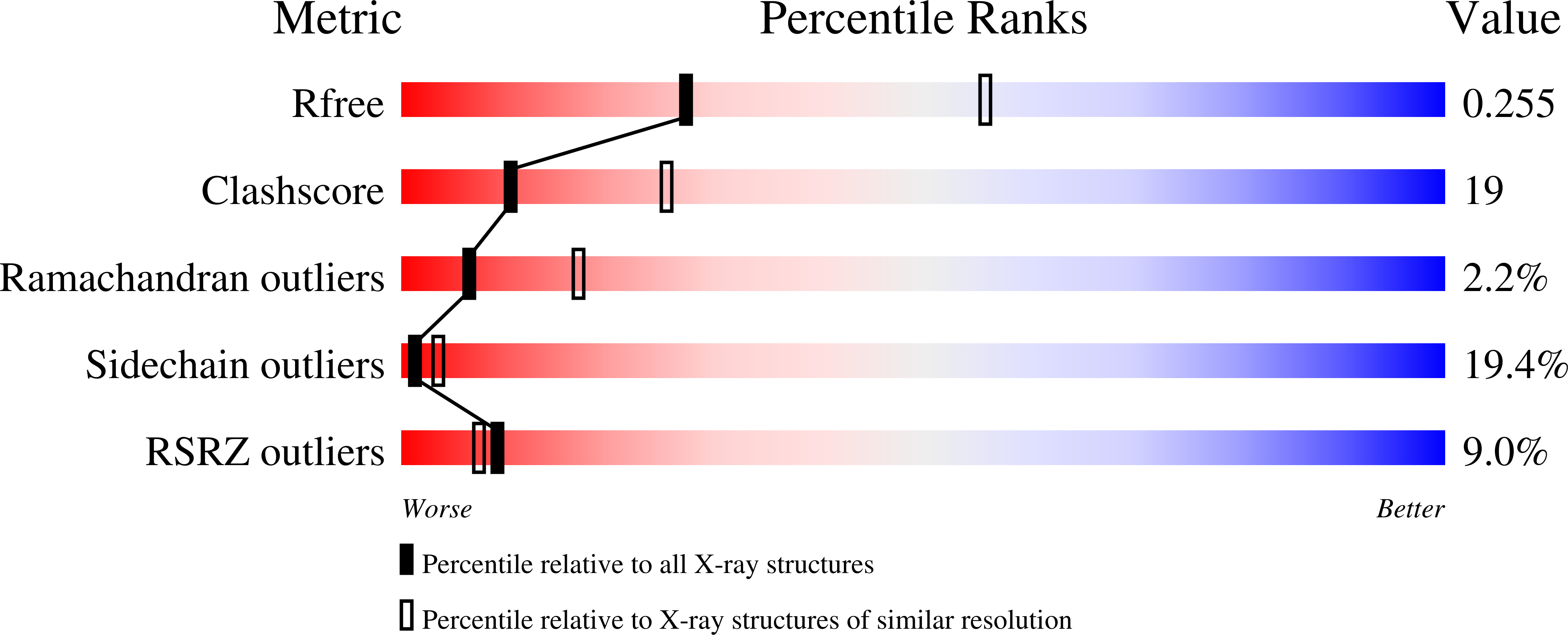
1S4D - PubMed Abstract:
The crystallographic structure of the Pseudomonas denitrificans S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase (SUMT), which is encoded by the cobA gene, has been solved by molecular replacement to 2.7A resolution. SUMT is a branchpoint enzyme that plays a key role in the biosynthesis of modified tetrapyrroles by controlling flux to compounds such as vitamin B(12) and sirohaem, and catalysing the transformation of uroporphyrinogen III into precorrin-2. The overall topology of the enzyme is similar to that of the SUMT module of sirohaem synthase (CysG) and the cobalt-precorrin-4 methyltransferase CbiF and, as with the latter structures, SUMT has the product S-adenosyl-L-homocysteine bound in the crystal. The roles of a number of residues within the SUMT structure are discussed with respect to their conservation either across the broader family of cobalamin biosynthetic methyltransferases or within the sub-group of SUMT members. The D47N, L49A, F106A, T130A, Y183A and M184A variants of SUMT were generated by mutagenesis of the cobA gene, and tested for SAM binding and enzymatic activity. Of these variants, only D47N and L49A bound the co-substrate S-adenosyl-L-methionine. Consequently, all the mutants were severely restricted in their capacity to synthesise precorrin-2, although both the D47N and L49A variants produced significant quantities of precorrin-1, the monomethylated derivative of uroporphyrinogen III. The activity of these variants is interpreted with respect to the structure of the enzyme.
Organizational Affiliation:
Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5YW, UK.