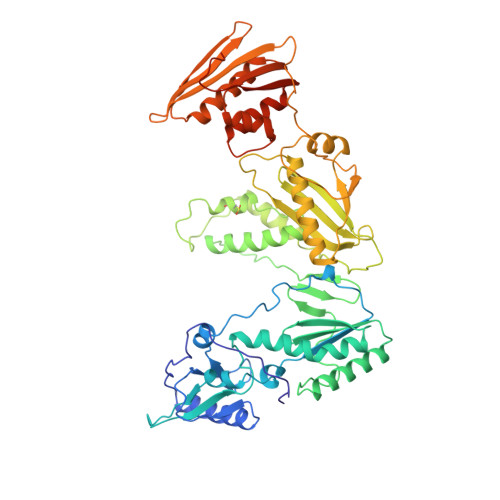
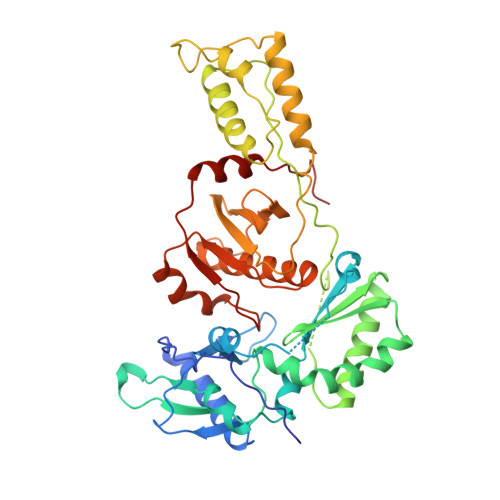
High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Ren, J., Esnouf, R., Garman, E., Somers, D., Ross, C., Kirby, I., Keeling, J., Darby, G., Jones, Y., Stuart, D.(1995) Nat Struct Biol 2: 293-302
- PubMed: 7540934
- DOI: https://doi.org/10.1038/nsb0495-293
- Primary Citation of Related Structures:
1RTH, 1RTI, 1VRT, 1VRU - PubMed Abstract:
We have determined the structures of four complexes of HIV-1 reverse transcriptase with non-nucleoside inhibitors, three fully refined at high resolution. The highest resolution structure is of the RT-nevirapine complex which has an R-factor of 0.186 and a root-mean-square bond length deviation of 0.015 A for all data to 2.2 A. The structures reveal a common mode of binding for these chemically diverse compounds. The common features of binding are largely hydrophobic interactions and arise from induced shape complementarity achieved by conformational rearrangement of the enzyme and conformational/configurational rearrangement of the compounds.
Organizational Affiliation:
Laboratory of Molecular Biophysics, Oxford, UK.