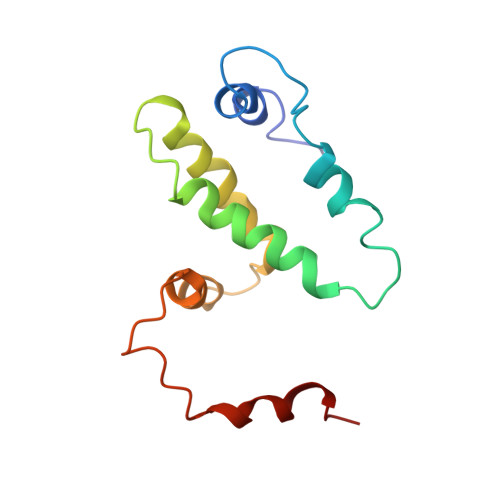
Structure of recombinant bovine interferon-gamma at 3.0 A resolution.
Samudzi, C.T., Rubin, J.R.(1993) Acta Crystallogr D Biol Crystallogr 49: 513-521
- PubMed: 15299487
- DOI: https://doi.org/10.1107/S0907444993006924
- Primary Citation of Related Structures:
1RFB - PubMed Abstract:
The three-dimensional crystal structure of recombinant bovine interferon-gamma was determined using the multiple isomorphous replacement method at 3.0 A and refined to an R factor of 19.2%. This protein crystallizes in space group P2(1)2(1)2(1) with unit-cell parameters of a = 42.8, b = 79.9 and c = 85.4 A. There is one functional dimer in the asymmetric unit. The two polypeptide chains are related by a non-crystallographic twofold symmetry axis. The secondary structure is predominantly alpha-helical with extensive interdigitation of the alpha-helical segments of the polypeptide chains that make up the dimer. The secondary structure, tertiary structure and topology of this molecule are identical to the previously reported structures of recombinant rabbit interferon-gamma and recombinant human interferon-gamma. The molecular topology is also similar to that of murine interferon-beta. These structural similarities strongly indicate the presence of a unique topological feature (fold) among gamma-interferons from different species, and also among the different classes of interferons.
Organizational Affiliation:
National Cancer Institute-Frederick Cancer Research Development Center, ABL-Basic Research Program, Frederick, Maryland 21702, USA.