Structure of Tandem RNA Recognition Motifs from Polypyrimidine Tract Binding Protein Reveals Novel Features of the Rrm Fold
Conte, M.R., Grune, T., Kelly, G., Ladas, A., Matthews, S., Curry, S.(2000) EMBO J 19: 3132
- PubMed: 10856256
- DOI: https://doi.org/10.1093/emboj/19.12.3132
- Primary Citation of Related Structures:
1QM9 - PubMed Abstract:
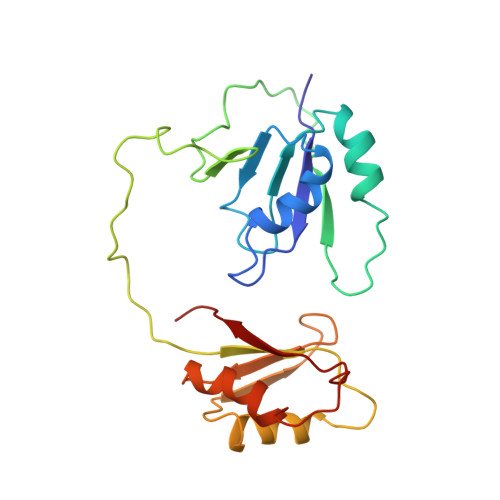
Polypyrimidine tract binding protein (PTB), an RNA binding protein containing four RNA recognition motifs (RRMs), is involved in both pre-mRNA splicing and translation initiation directed by picornaviral internal ribosome entry sites. Sequence comparisons previously indicated that PTB is a non-canonical RRM protein. The solution structure of a PTB fragment containing RRMs 3 and 4 shows that the protein consists of two domains connected by a long, flexible linker. The two domains tumble independently in solution, having no fixed relative orientation. In addition to the betaalphabetabetaalphabeta topology, which is characteristic of RRM domains, the C-terminal extension of PTB RRM-3 incorporates an unanticipated fifth beta-strand, which extends the RNA binding surface. The long, disordered polypeptide connecting beta4 and beta5 in RRM-3 is poised above the RNA binding surface and is likely to contribute to RNA recognition. Mutational analyses show that both RRM-3 and RRM-4 contribute to RNA binding specificity and that, despite its unusual sequence, PTB binds RNA in a manner akin to that of other RRM proteins.
Organizational Affiliation:
Department of Biochemistry, Imperial College of Science, Technology and Medicine, Exhibition Road, London, UK.