Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element
Jacks, A., Babon, J., Kelly, G., Manolaridis, I., Cary, P.D., Curry, S., Conte, M.R.(2003) Structure 11: 833-843
- PubMed: 12842046
- DOI: https://doi.org/10.1016/s0969-2126(03)00121-7
- Primary Citation of Related Structures:
1OWX - PubMed Abstract:
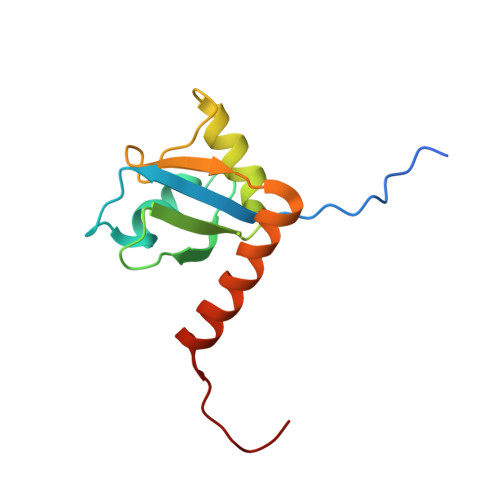
The La protein is an important component of ribonucleoprotein complexes that acts mainly as an RNA chaperone to facilitate correct processing and maturation of RNA polymerase III transcripts, but can also stimulate translation initiation. We report here the structure of the C-terminal domain of human La, which comprises an atypical RNA recognition motif (La225-334) and a long unstructured C-terminal tail. The central beta sheet of La225-334 reveals novel features: the putative RNA binding surface is formed by a five-stranded beta sheet and, strikingly, is largely obscured by a long C-terminal alpha helix that encompasses a recently identified nuclear retention element. Contrary to previous observations, we find that the La protein does not contain a dimerization domain.
Organizational Affiliation:
Biophysics Laboratories, Institute of Biomedical and Biomolecular Sciences, University of Portsmouth, St Michael's Building, PO1 2DT, Portsmouth, United.