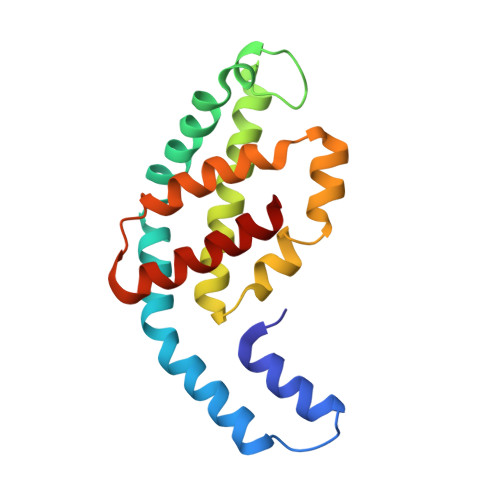
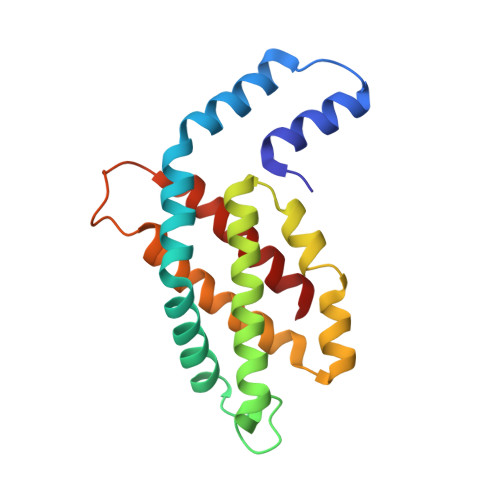
The crystal structure of a novel unmethylated form of C-phycocyanin, a possible connector between cores and rods in pycobilisomes
Adir, N., Lerner, N.(2003) J Biol Chem 278: 25926-25932
- PubMed: 12709431
- DOI: https://doi.org/10.1074/jbc.M302838200
- Primary Citation of Related Structures:
1ON7 - PubMed Abstract:
A novel fraction of c-phycocyanin from the thermophilic cyanobacterium Thermosynechcoccus vulcanus, with an absorption maxima blue-shifted to 612 nm (PC612), has been purified from allophycocyanin and crystallized. The crystals belong to the P63 space group with cell dimensions of 153 A x 153 A x 59 A with a single (alphabeta) monomer in the asymmetric unit, resulting in a solvent content of 65%, and diffract to 2.7 A. The PC612 crystal structure has been determined by molecular replacement and refined to a crystallographic R-factor of 20.9% (Rfree = 27.8%). The crystal packing in this form shows that the PC612 form of phycocyanin does not associate into hexamers and that its association with adjacent trimers in the unit cell is very different from that found in a previously determined structure of the normal form of T. vulcanus phycocyanin, which absorbs at 620 nm. Analysis of the PC612 structure shows that the alpha subunits, which typically form the interface between two trimers within a hexamer, have a high degree of flexibility, as indicated by elevated B-factors in portions of helices B, E, and G. Examination of calculated electron density omit maps shows that unlike all other structures of phycobiliproteins determined so far, the Asnbeta72 residue is not methylated, explaining the blue-shift in its absorption spectra. On the basis of the results presented here, we suggest that this new form of trimeric phycocyanin may constitute a special minor component of the phycobilisome and may form the contact between the phycocyanin rods and the allophycocyanin core.
Organizational Affiliation:
Department of Chemistry and Institute of Catalysis, Science and Technology, Technion, Israel Institute of Technology, Technion City, Haifa 32000, Israel.