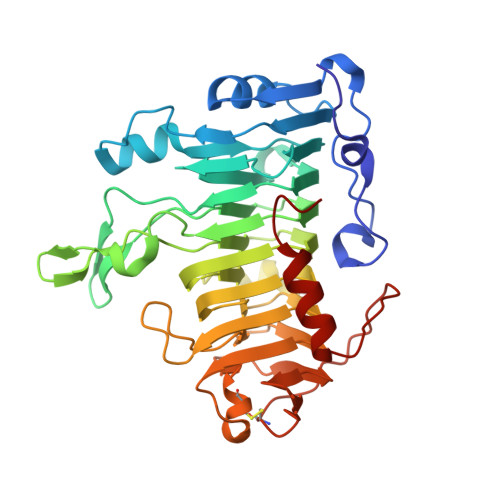
Structure of pectate lyase A: comparison to other isoforms.
Thomas, L.M., Doan, C.N., Oliver, R.L., Yoder, M.D.(2002) Acta Crystallogr D Biol Crystallogr 58: 1008-1015
- PubMed: 12037303
- DOI: https://doi.org/10.1107/s0907444902005851
- Primary Citation of Related Structures:
1JRG, 1JTA - PubMed Abstract:
Pectate lyase A is a virulence factor secreted by the plant-pathogenic bacteria Erwinia chrysanthemi. The enzyme cleaves the glycosidic bond of pectate polymers by a calcium-dependent beta-elimination mechanism. The crystal structure of pectate lyase A from E. chrysanthemi EC16 has been determined in two crystal forms, monoclinic C2 to 1.8 A and rhombohedral R3 to 2.1 A. The protein structure is compared with two other pectate lyase isoforms from E. chrysanthemi EC16, pectate lyase C and pectate lyase E. Pectate lyase A is unique as it is the only acidic pectate lyase and has end products that are significantly more varied in length in comparison to those of the other four major pectate lyase isozymes. Differences and similarities in polypeptide trace, size and volume of the active-site groove and surface electrostatics are discussed.
Organizational Affiliation:
School of Biological Sciences, University of Missouri-Kansas City, 5007 Rockhill Road, Kansas City, MO 64110-2499, USA.