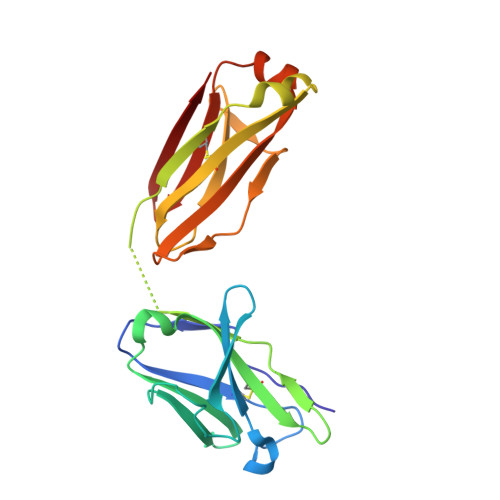
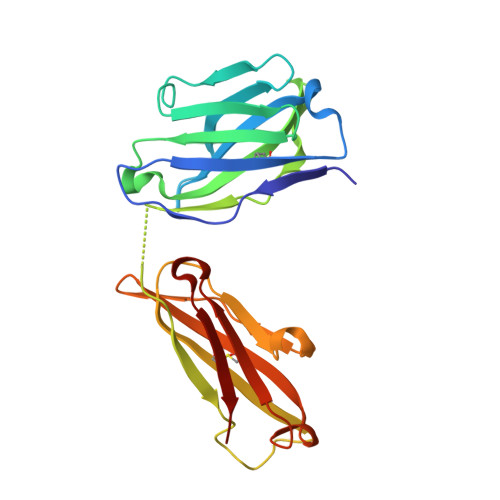
Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
Monnet, C., Bettsworth, F., Stura, E.A., Du, M.H., Menez, R., Derrien, L., Zinn-Justin, S., Gilquin, B., Sibai, G., Battail-Poirot, N., Jolivet, M., Menez, A., Arnaud, M., Ducancel, F., Charbonnier, J.B.(2002) J Mol Biol 315: 699-712
- PubMed: 11812141
- DOI: https://doi.org/10.1006/jmbi.2001.5284
- Primary Citation of Related Structures:
1JN6, 1JNH, 1JNL, 1JNN - PubMed Abstract:
Subtle modulation of antibody-binding properties by protein engineering often lies with an accurate structural and energetic description of how an antigen is recognised. Thus, with the intent to increase the affinity and add a bias in favour of natural estradiol compared with its chemically modified immunogen, we have determined the crystal structure of two anti-estradiol monoclonal antibodies, 10G6D6 and 17E12E5. Although generated against the same estradiol derivative, these antibodies share little sequence identity, which is reflected in dissimilar binding pockets and in different positioning of the steroid. In both antibodies the characteristic 17-hydroxyl group is buried deeply at the bottom of hydrophobic pockets and stabilised by hydrogen bonds. Apart from this similarity, the steroid is oriented differently in the respective binding pockets. The high specificity of both antibodies has been mapped out, and even closely related steroids show low cross-reactivity. The structural studies of the complex formed between 10G6D6 and 6-CMO-estradiol have identified contacts between the 6-CMO coupling linker and an arginine residue from the heavy chain CDR2 segment. This segment is now being targeted by random mutagenesis to select mutants with a preference for natural estradiol compared to the branched hapten.
Organizational Affiliation:
Département d'Ingénierie et d'Etude des Protéines, CEA, CE Saclay, Gif-sur-Yvette Cedex, 91191, France.