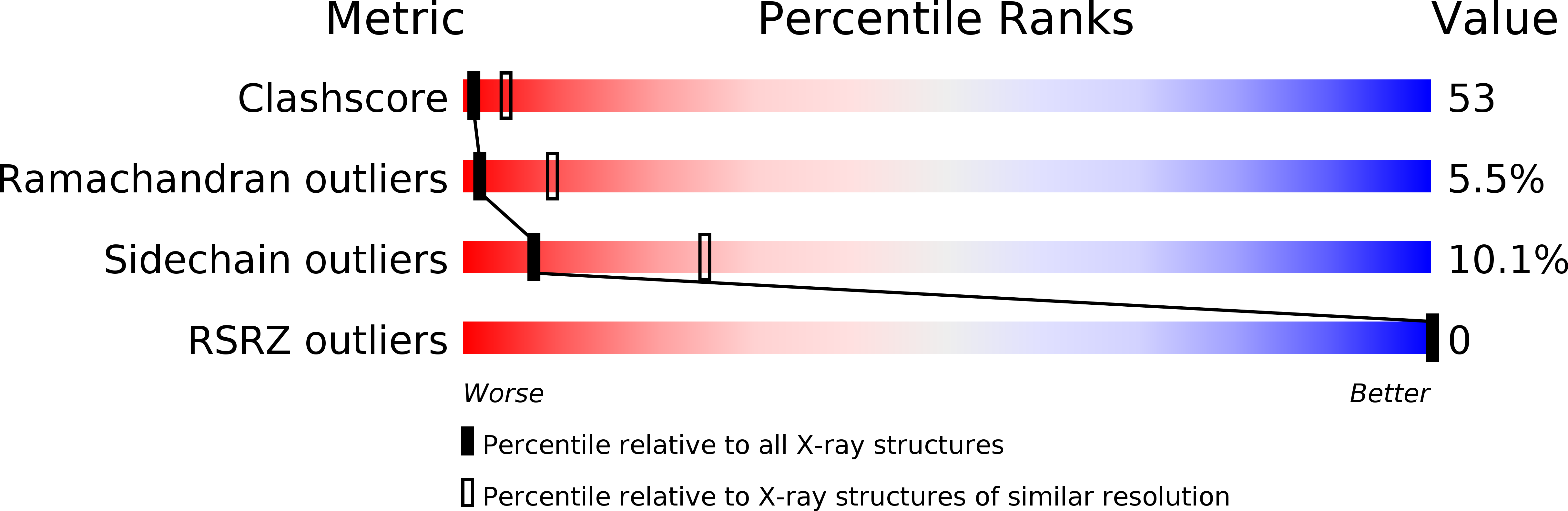
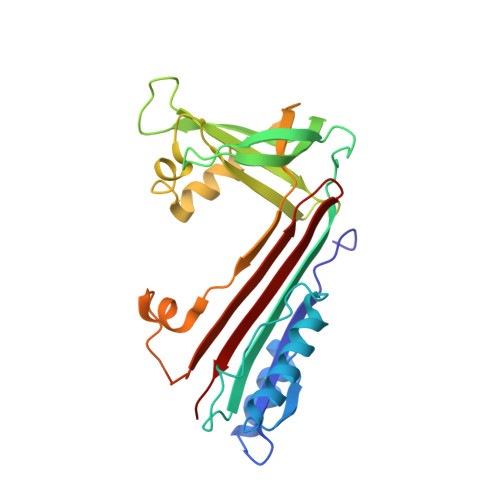
Crystal structure of neuroserpin: a neuronal serpin involved in a conformational disease.
Briand, C., Kozlov, S.V., Sonderegger, P., Grutter, M.G.(2001) FEBS Lett 505: 18-22
- PubMed: 11557034
- DOI: https://doi.org/10.1016/s0014-5793(01)02764-8
- Primary Citation of Related Structures:
1JJO - PubMed Abstract:
The protease inhibitor neuroserpin regulates the development of the nervous system and its plasticity in the adult. Neuroserpins carrying the Ser53Pro or Ser56Arg mutation form polymers in neuronal cells. We describe here the structure of wild-type neuroserpin in a cleaved form. The structure provides a basis to understand the role of the mutations in the polymerization process. We propose that these mutations could delay the insertion of the reactive center loop into the central beta-sheet A, an essential step in the inhibition and possibly in the polymerization of neuroserpin.
Organizational Affiliation:
Institute of Biochemistry, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland.