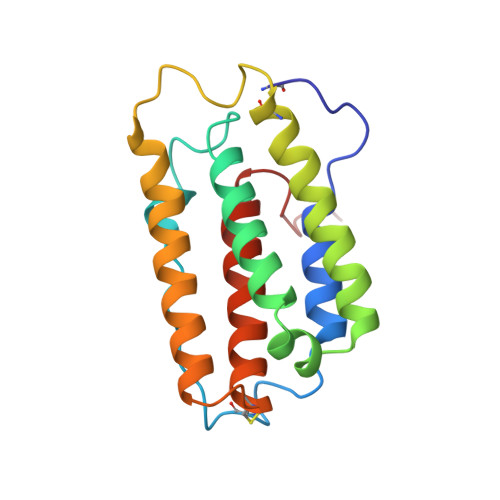
The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
Klaus, W., Gsell, B., Labhardt, A.M., Wipf, B., Senn, H.(1997) J Mol Biol 274: 661-675
- PubMed: 9417943
- DOI: https://doi.org/10.1006/jmbi.1997.1396
- Primary Citation of Related Structures:
1ITF - PubMed Abstract:
The solution structure of recombinant human interferon alpha-2a (Roferon-A) has been determined by multidimensional heteronuclear NMR spectroscopy. The calculations using simulated annealing produced a family of 24 convergent structures which satisfy the experimental restraints comprising 1541 NOE-derived inter-proton distances, 187 dihedral restraints, 66 pairs of hydrogen bond restraints, and six upper and lower limits for two disulfide bridges. The fractional labeling of methyl groups allowed their direct and unambiguous stereospecific assignment which proved to be essential for obtaining a high resolution of the structures. A best fit superposition of residues 10 to 47, 50 to 101 and 111 to 157 gives an rms deviation of 0.62 A for the backbone heavy atoms and 1.39 A for all heavy atoms of these segments. The dominant feature of the structure is a cluster of five alpha-helices, four of which are arranged to form a left-handed helix bundle with an up-up-down-down topology and two over-hand connections. The interpretation of heteronuclear 15N-¿1H¿ NOE data shows the co-existence of flexible regions within an otherwise rigid framework of the protein. Four stretches of pronounced flexibility can be located: Cys1-Ser8, Gly44-Ala50, Ile100-Lys112, and Ser160-Glu165. Among the structurally related four-helical bundle cytokines, the structure of IFN alpha-2a is most similar to that of human interferon alpha-2b and murine interferon-beta. From this structural information and mutagenesis data, areas on the surface of the protein are identified which seem to be important in receptor interactions.
Organizational Affiliation:
F. Hoffmann-LaRoche AG Pharma Preclinical Research Department, Basel, Switzerland.