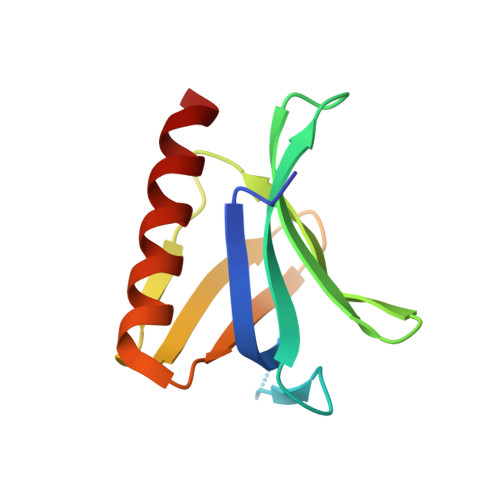
The N-terminal domain of Homer/Vesl is a new class II EVH1 domain.
Barzik, M., Carl, U.D., Schubert, W.D., Frank, R., Wehland, J., Heinz, D.W.(2001) J Mol Biol 309: 155-169
- PubMed: 11491285
- DOI: https://doi.org/10.1006/jmbi.2001.4640
- Primary Citation of Related Structures:
1I7A - PubMed Abstract:
Cellular activities controlled by signal transduction processes such as cell motility and cell growth depend on the tightly regulated assembly of multiprotein complexes. Adapter proteins that specifically interact with their target proteins are key components required for the formation of these assemblies. Ena/VASP-homology 1 (EVH1) domains are small constituents of large modular proteins involved in microfilament assembly that specifically recognize proline-rich regions. EVH1 domain-containing proteins are present in neuronal cells, like the Homer/Vesl protein family that is involved in memory-generating processes. Here, we describe the crystal structure of the murine EVH1 domain of Vesl 2 at 2.2 A resolution. The small globular protein consists of a seven-stranded antiparallel beta-barrel with a C-terminal alpha-helix packing alongside the barrel. A shallow groove running parallel with beta-strand VI forms an extended peptide-binding site. Using peptide library screenings, we present data that demonstrate the high affinity of the Vesl 2 EVH1 domain towards peptide sequences containing a proline-rich core sequence (PPSPF) that requires additional charged amino acid residues on either side for specific binding. Our functional data, substantiated by structural data, demonstrate that the ligand-binding of the Vesl EVH1 domain differs from the interaction characteristics of the previously examined EVH1 domains of the Evl/Mena proteins. Analogous to the Src homology 3 (SH3) domains that bind their cognate ligands in two distinct directions, we therefore propose the existence of two distinct classes of EVH1 domains.
Organizational Affiliation:
Department of Structural Biology and German National Center of Biotechnology (GBF), Braunschweig.