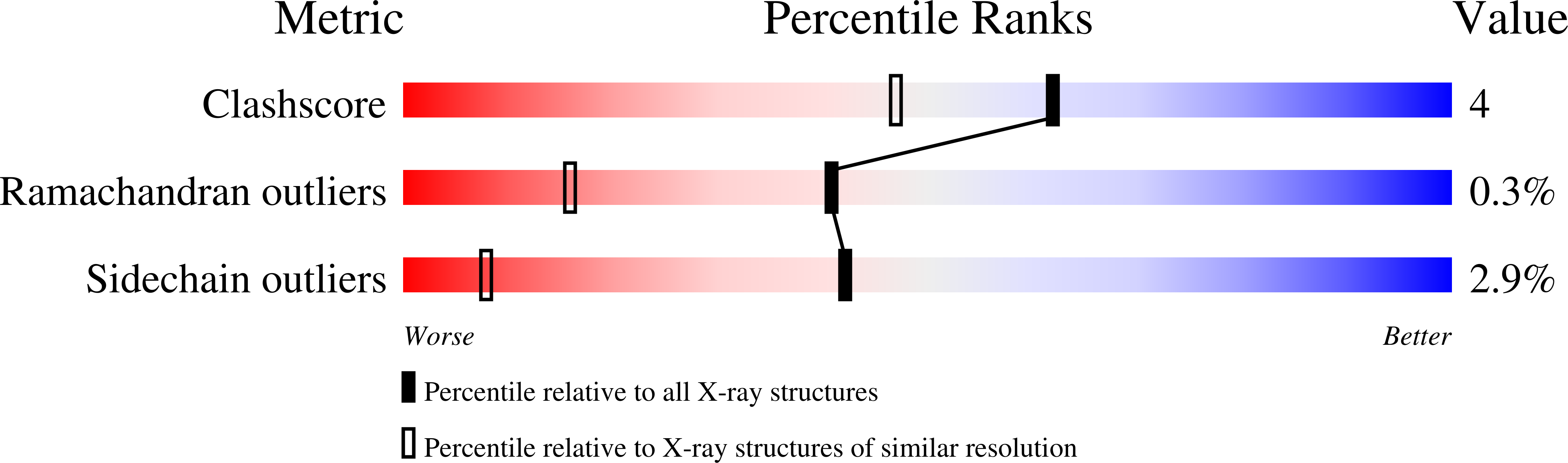
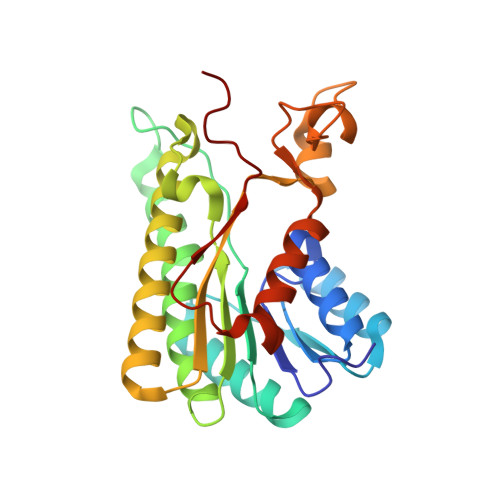
Structure of Bacterial 3beta/17beta-Hydroxysteroid Dehydrogenase at 1.2 A Resolution: A Model for Multiple Steroid Recognition
Benach, J., Filling, C., Oppermann, U.C.T., Roversi, P., Bricogne, G., Berndt, K.D., Jornvall, H., Ladenstein, R.(2002) Biochemistry 41: 14659-14668
- PubMed: 12475215
- DOI: https://doi.org/10.1021/bi0203684
- Primary Citation of Related Structures:
1HXH - PubMed Abstract:
The enzyme 3beta/17beta-hydroxysteroid dehydrogenase (3beta/17beta-HSD) is a steroid-inducible component of the Gram-negative bacterium Comamonas testosteroni. It catalyzes the reversible reduction/dehydrogenation of the oxo/beta-hydroxy groups at positions 3 and 17 of steroid compounds, including hormones and isobile acids. Crystallographic analysis at 1.2 A resolution reveals the enzyme to have nearly identical subunits that form a tetramer with 222 symmetry. This is one of the largest oligomeric structures refined at this resolution. The subunit consists of a monomer with a single-domain structure built around a seven-stranded beta-sheet flanked by six alpha-helices. The active site contains a Ser-Tyr-Lys triad, typical for short-chain dehydrogenases/reductases (SDR). Despite their highly diverse substrate specificities, SDR members show a close to identical folding pattern architectures and a common catalytic mechanism. In contrast to other SDR apostructures determined, the substrate binding loop is well-defined. Analysis of structure-activity relationships of catalytic cleft residues, docking analysis of substrates and inhibitors, and accessible surface analysis explains how 3beta/17beta-HSD accommodates steroid substrates of different conformations.
Organizational Affiliation:
Karolinska Institutet NOVUM, Center for Structural Biochemistry, S-14157 Huddinge, Sweden.