A STRUCTURAL EXPLANATION FOR ENZYME MEMORY IN NONAQUEOUS SOLVENTS.
Yennawar, H.P., Yennawar, N.H., Farber, G.K.(1995) J Am Chem Soc 117: 577-585
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(1995) J Am Chem Soc 117: 577-585
Find similar proteins by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GAMMA-CHYMOTRYPSIN | A [auth E] | 13 | Bos taurus | Mutation(s): 0 EC: 3.4.21.1 |  |
UniProt | |||||
Find proteins for P00766 (Bos taurus) Explore P00766 Go to UniProtKB: P00766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GAMMA-CHYMOTRYPSIN | B [auth F] | 131 | Bos taurus | Mutation(s): 0 EC: 3.4.21.1 | 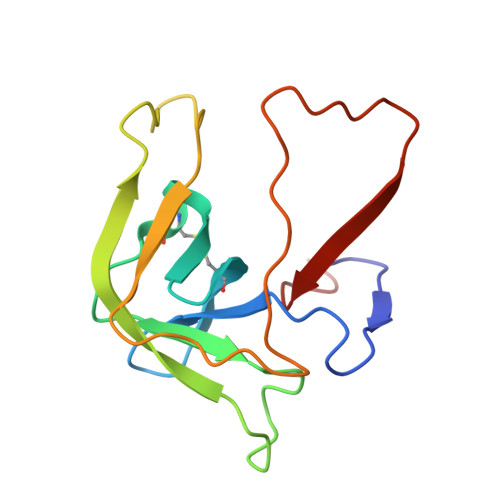 |
UniProt | |||||
Find proteins for P00766 (Bos taurus) Explore P00766 Go to UniProtKB: P00766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GAMMA-CHYMOTRYPSIN | C [auth G] | 97 | Bos taurus | Mutation(s): 0 EC: 3.4.21.1 | 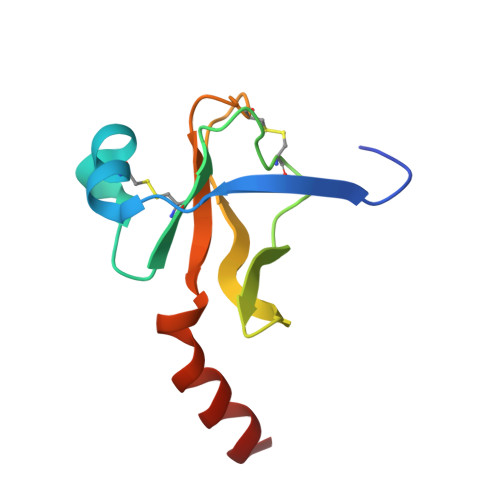 |
UniProt | |||||
Find proteins for P00766 (Bos taurus) Explore P00766 Go to UniProtKB: P00766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00766 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PRO-GLY-ALA | D [auth P] | 3 | N/A | Mutation(s): 0 | 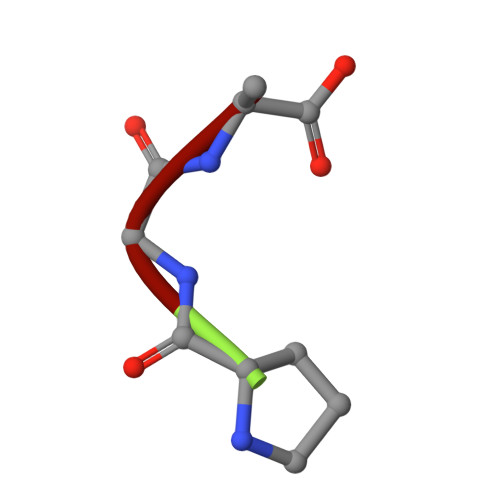 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| TRP Query on TRP | F [auth E] | TRYPTOPHAN C11 H12 N2 O2 QIVBCDIJIAJPQS-VIFPVBQESA-N |  | ||
| HEX Query on HEX | G [auth E], H [auth E], I [auth F], J [auth F], L [auth G] | HEXANE C6 H14 VLKZOEOYAKHREP-UHFFFAOYSA-N |  | ||
| IPA Query on IPA | K [auth F], M [auth G], N [auth G] | ISOPROPYL ALCOHOL C3 H8 O KFZMGEQAYNKOFK-UHFFFAOYSA-N |  | ||
| ACE Query on ACE | E | ACETYL GROUP C2 H4 O IKHGUXGNUITLKF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.6 | α = 90 |
| b = 69.6 | β = 90 |
| c = 97.51 | γ = 90 |
| Software Name | Purpose |
|---|---|
| X-PLOR | model building |
| X-PLOR | refinement |
| X-PLOR | phasing |