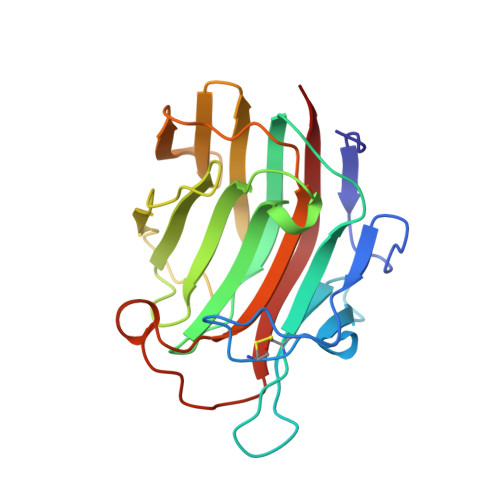
Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
Hahn, M., Pons, J., Planas, A., Querol, E., Heinemann, U.(1995) FEBS Lett 374: 221-224
- PubMed: 7589539
- DOI: https://doi.org/10.1016/0014-5793(95)01111-q
- Primary Citation of Related Structures:
1GBG - PubMed Abstract:
The crystal structure of the 1,3-1,4-beta-D-glucan 4-glucanohydrolase from Bacillus licheniformis is solved at a resolution of 1.8 A and refined to R = 16.5%. The protein has a similar beta-sandwich structure as the homologous enzyme from Bacillus macerans and the hybrid H(A16-M). This demonstrates that the jellyroll fold of these proteins is remarkably rigid and only weakly influenced by crystal contacts. The crystal structure permits to extend mechanistic considerations derived for the B. licheniformis enzyme to the entire class of bacterial 1,3-1,4-beta-D-glucan 4-glucanohydrolases.
Organizational Affiliation:
Forschungsgruppe Kristallographie, Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany.