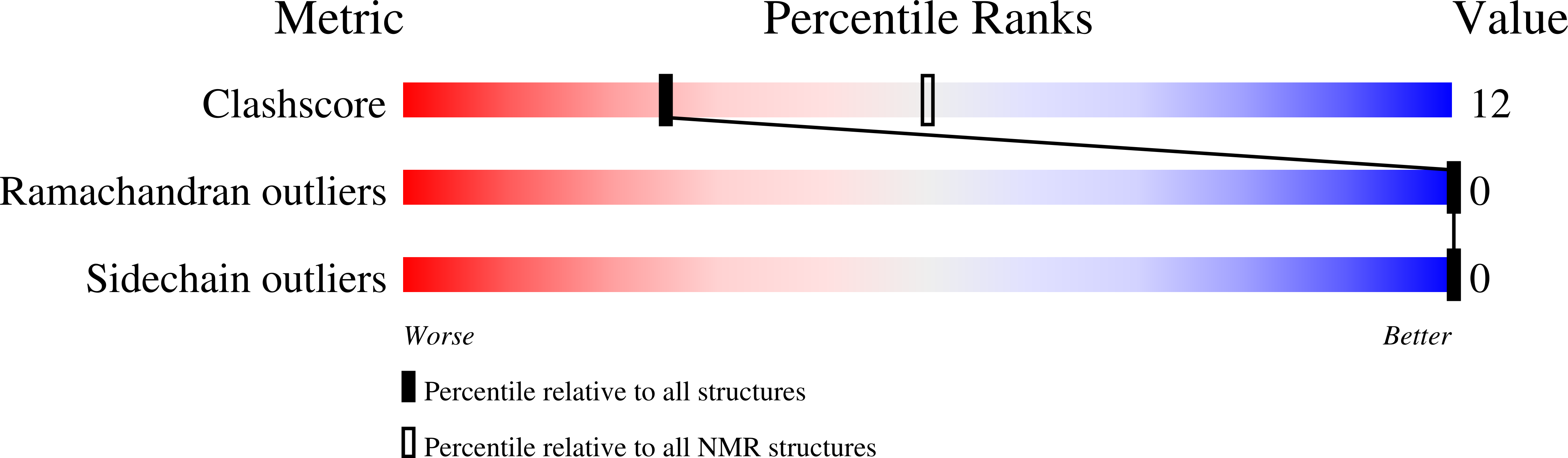DNA Bending and Unwinding Associated with Actinomycin D Antibiotics Bound to Partially Overlapping Sites on DNA.
Chen, H., Liu, X., Patel, D.J.(1996) J Mol Biol 258: 457
- PubMed: 8642603
- DOI: https://doi.org/10.1006/jmbi.1996.0262
- Primary Citation of Related Structures:
1FJA - PubMed Abstract:
Actinomycin D (ActD) is a potent anti-tumor antibiotic, that preferentially targets (G-C).(G-C) steps on duplex DNA. We have reported on the solution structure of the ActD-d(A-A-A-G-C-T-T-T) complex (one drug per duplex) based on a combined application of NMR and molecular dynamics calculations. This study established that ActD binds to DNA through intercalation of the phenoxazone chromophore between (G-C).(G-C) steps with the benzenoid and quinonoid-linked cyclic pentapeptide lactone rings spanning two base-pairs in opposite directions in the minor groove of the helix. This research is now extended to the binding of two ActD molecules to adjacent complexation sites within a (G-C-G-C).(G-C-G-C) segment in the self-complementary d(A1-A2-G3-C4-G5-C6-T7-T8) duplex. The occupation of the central (C4-G5).(C4-G5) segment between the two intercalation sites by the inwardly pointing cyclic pentapeptide lactone rings from adjacent bound ActD molecules should result in a potential steric clash in the center of the helix. The NMR data and its analysis on the ActD-d(A-A-G-C-G-C-T-T) complex (two drugs per duplex) establish that two ActD molecules intercalate into symmetry-related (G3-C4).(G5-C6) steps with their attached benzenoid and quinonoid cyclic pentapeptide lactone rings positioned in the minor groove and directed towards the center and the ends of the helix, respectively. The solution structure of the complex was solved by using NMR restraints to guide distance geometry-simulated annealing and restrained molecular dynamics calculations including intensity-based refinements. The DNA helix exhibits a pronounced kink and is fully unwound at the central (C4-G5).(C4-G5) step which results in an opening and widening of the minor groove to generate additional space for accommodation of the inwardly pointing benzenoid cyclic pentapeptide lactone rings in the complex. The outwardly and inwardly pointing cyclic pentapeptide lactone rings of symmetry-related ActD molecules retain similar conformations with the largest difference observed for the L-MeVal residues in the complex. The present study defines how structural changes primarily in the DNA associated with the directional bending of the helix towards the major groove and away from the bound drug opens up and widens the minor groove to accommodate two intercalated ActD molecules bound at partially overlapping sites on the DNA.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, College of Physicians and Surgeons of Columbia University, New York, NY 10032, USA.