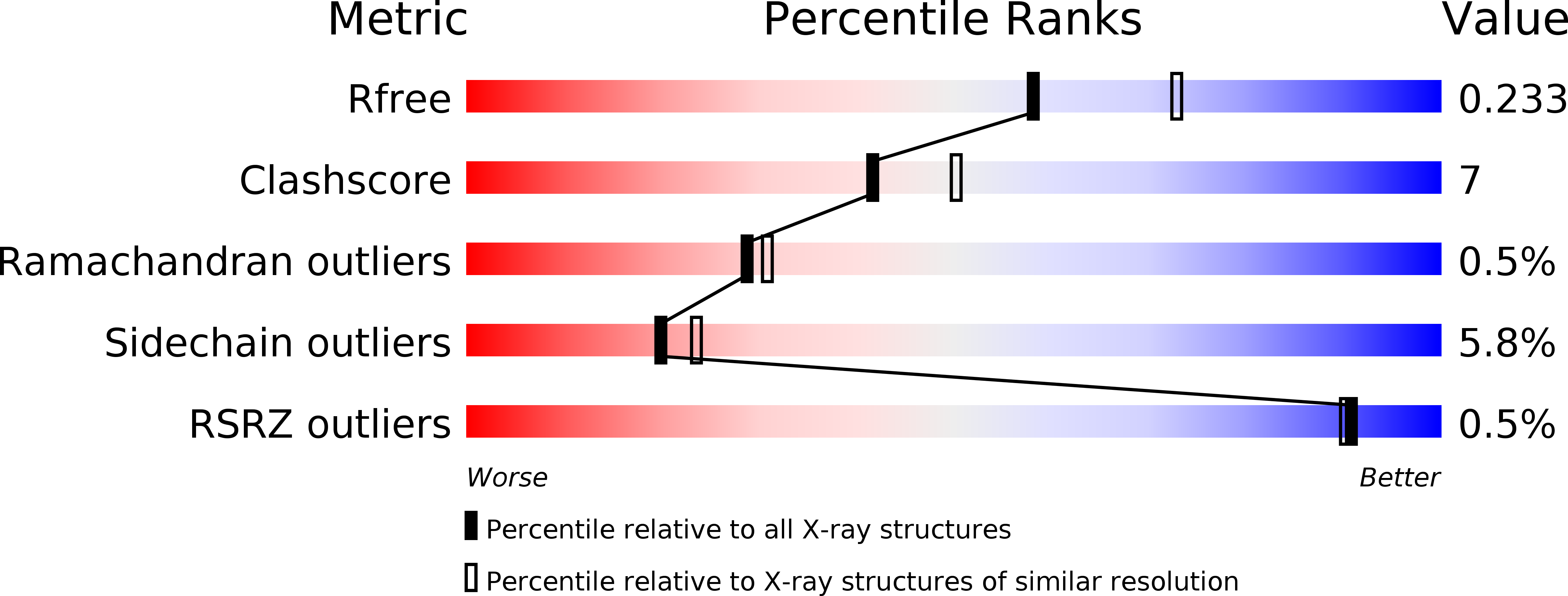
Crystal structure of the dihaem cytochrome c4 from Pseudomonas stutzeri determined at 2.2A resolution.
Kadziola, A., Larsen, S.(1997) Structure 5: 203-216
- PubMed: 9032080
- DOI: https://doi.org/10.1016/s0969-2126(97)00179-2
- Primary Citation of Related Structures:
1ETP - PubMed Abstract:
. Cytochromes c4 are dihaem cytochromes c found in a variety of bacteria. They are assumed to take part in the electron-transport systems associated with both aerobic and anaerobic respiration. The cytochrome c4 proteins are located in the periplasm, predominantly bound to the inner membrane, and are able to transfer electrons between membrane-bound reduction systems and terminal oxidases. Alignment of cytochrome c4 sequences from three bacteria, Pseudomonas aeruginosa, Pseudomonas stutzeri and Azotobacter vinelandii, suggests that these dihaem proteins are composed of two similar domains. Two distinctly different redox potentials have been measured for the Ps. stutzeri cytochrome c4, however. . The crystal structure of the dihaem cytochrome c4 from Ps. stutzeri has been determined to 2.2A resolution by isomorphous replacement. The model, consisting of two entire cytochrome c4 molecules and 138 water molecules in the asymmetric unit, was refined to an R value of 20.1% for all observations in the resolution range 8-2.2A. The molecule is organized in two cytochrome c-like domains that are related by a pseudo-twofold axis. The symmetry is virtually perfectly close to the twofold axis, which passes through a short hydrogen bond between the two haem propionic acid groups, connecting the redox centre of each domain. This haem-haem interaction is further stabilized by an extensive symmetrical hydrogen-bond network. The twofold symmetry is not present further away from the axis, however, and the cytochrome c4 molecule can be considered to be a dipole with charged residues unevenly distributed between the two domains. The haem environment in the two domains show pronounced differences, mainly on the methionine side of the haem group. . The structure, in conjunction with sequence alignment, suggests that the cytochrome protein has evolved by duplication of a cytochrome c gene. The difference in charge distribution around each haem group in the two domains allows the haem group in the N-terminal domain to be associated with the lower redox potential of 241 mV and the C-terminal haem group with the higher potential of 328 mV. The molecular dipole characteristic of cytochrome c4 is important for its interaction with, and recognition of, its redox partners. In cytochrome c4, the hydrogen-bond network (between residues that are conserved in all known cytochrome c4 subspecies) seems to provide an efficient pathway for an intramolecular electron transfer that can ensure cooperativity between the two redox centres. The C-pyrrole corners of the haem edges are potential sites for external electron exchange.
Organizational Affiliation:
Centre for Crystallographic Studies, Department of Chemistry, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen, Denmark.