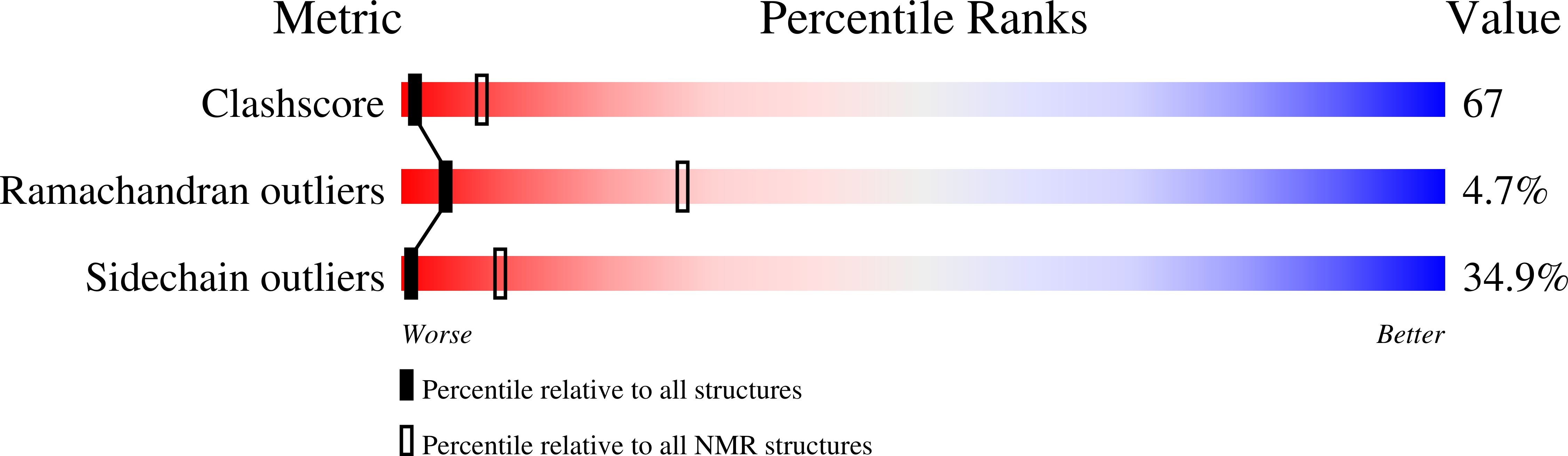Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70
Booth, V., Koth, C., Edwards, A.M., Arrowsmith, C.H.(2000) J Biol Chem 275: 31266-31268
- PubMed: 10811649
- DOI: https://doi.org/10.1074/jbc.M002595200
- Primary Citation of Related Structures:
1EO0 - PubMed Abstract:
TFIIS is a transcription elongation factor that consists of three domains. We have previously solved the structures of domains II and III, which stimulate arrested polymerase II elongation complexes in order to resume transcription. Domain I is conserved in evolution from yeast to human species and is homologous to the transcription factors elongin A and CRSP70. Domain I also interacts with the transcriptionally active RNA polymerase II holoenzyme and therefore, may have a function unrelated to the previously described transcription elongation activity of TFIIS. We have solved the structure of domain I of yeast TFIIS using NMR spectroscopy. Domain I is a compact four-helix bundle that is structurally independent of domains II and III of the TFIIS. Using the yeast structure as a template, we have modeled the homologous domains from elongin A and CRSP70 and identified a conserved positively charged patch on the surface of all three proteins, which may be involved in conserved functional interactions with the transcriptional machinery.
Organizational Affiliation:
Ontario Cancer Institute and Department of Medical Biophysics, University of Toronto, Toronto, Ontario M5G 2M9, Canada.