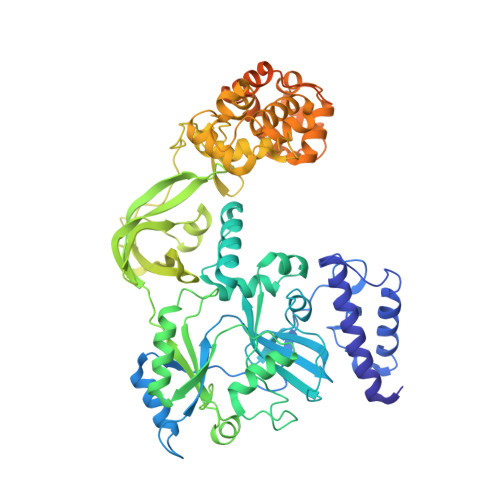
Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Lee, J.Y., Chang, C., Song, H.K., Moon, J., Yang, J.K., Kim, H.K., Kwon, S.T., Suh, S.W.(2000) EMBO J 19: 1119-1129
- PubMed: 10698952
- DOI: https://doi.org/10.1093/emboj/19.5.1119
- Primary Citation of Related Structures:
1DGS, 1V9P - PubMed Abstract:
DNA ligases catalyze the crucial step of joining the breaks in duplex DNA during DNA replication, repair and recombination, utilizing either ATP or NAD(+) as a cofactor. Despite the difference in cofactor specificity and limited overall sequence similarity, the two classes of DNA ligase share basically the same catalytic mechanism. In this study, the crystal structure of an NAD(+)-dependent DNA ligase from Thermus filiformis, a 667 residue multidomain protein, has been determined by the multiwavelength anomalous diffraction (MAD) method. It reveals highly modular architecture and a unique circular arrangement of its four distinct domains. It also provides clues for protein flexibility and DNA-binding sites. A model for the multidomain ligase action involving large conformational changes is proposed.
Organizational Affiliation:
Center for Molecular Catalysis, Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul 151-742.