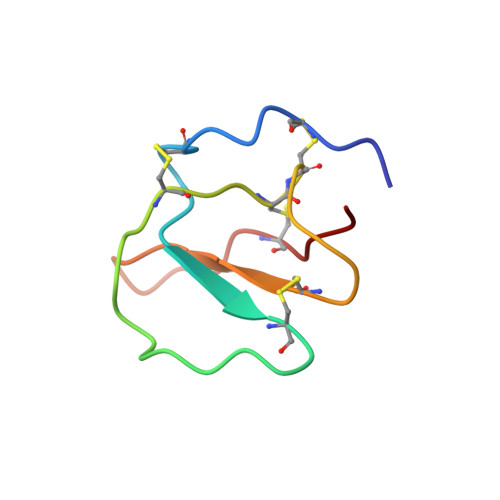
Structure of a putative ancestral protein encoded by a single sequence repeat from a multidomain proteinase inhibitor gene from Nicotiana alata.
Scanlon, M.J., Lee, M.C., Anderson, M.A., Craik, D.J.(1999) Structure 7: 793-802
- PubMed: 10425681
- DOI: https://doi.org/10.1016/s0969-2126(99)80103-8
- Primary Citation of Related Structures:
1CE3 - PubMed Abstract:
The ornamental tobacco Nicotiana alata produces a series of proteinase inhibitors (PIs) that are derived from a 43 kDa precursor protein, NaProPI. NaProPI contains six highly homologous repeats that fold to generate six separate structural domains, each corresponding to one of the native PIs. An unusual feature of NaProPI is that the structural domains lie across adjacent repeats and that the sixth PI domain is generated from fragments of the first and sixth repeats. Although the homology of the repeats suggests that they may have arisen from gene duplication, the observed folding does not appear to support this. This study of the solution structure of a single NaProPI repeat (aPI1) forms a basis for unravelling the mechanism by which this protein may have evolved. The three-dimensional structure of aPI1 closely resembles the triple-stranded antiparallel beta sheet observed in each of the native PIs. The five-residue sequence Glu-Glu-Lys-Lys-Asn, which forms the linker between the six structural domains in NaProPI, exists as a disordered loop in aPI1. The presence of this loop in aPI1 results in a loss of the characteristically flat and disc-like topography of the native inhibitors. A single repeat from NaProPI is capable of folding into a compact globular domain that displays native-like PI activity. Consequently, it is possible that a similar single-domain inhibitor represents the ancestral protein from which NaProPI evolved.
Organizational Affiliation:
Centre for Drug Design and Development, University of Queensland, St Lucia, Australia.