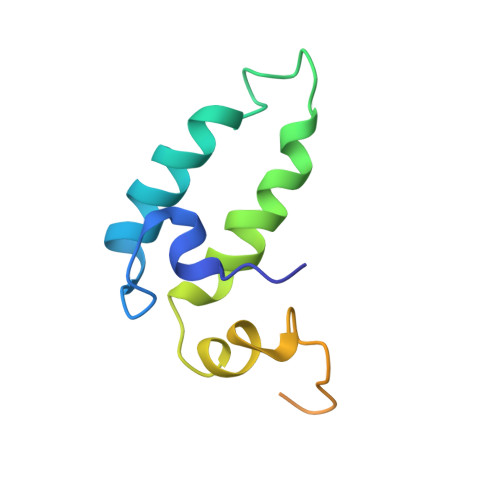
The influence of C-terminal extension on the structure of the "J-domain" in E. coli DnaJ.
Huang, K., Flanagan, J.M., Prestegard, J.H.(1999) Protein Sci 8: 203-214
- PubMed: 10210198
- DOI: https://doi.org/10.1110/ps.8.1.203
- Primary Citation of Related Structures:
1BQ0, 1BQZ - PubMed Abstract:
Two different recombinant constructs of the N-terminal domain in Escherichia coli DnaJ were uniformly labeled with nitrogen-15 and carbon-13. One, DnaJ(1-78), contains the complete "J-domain," and the other, DnaJ(1-104), contains both the "J-domain" and a conserved "G/F" extension at the C-terminus. The three-dimensional structures of these proteins have been determined by heteronuclear NMR experiments. In both proteins the "J-domain" adopts a compact structure consisting of a helix-turn-helix-loop-helix-turn-helix motif. In contrast, the "G/F" region in DnaJ(1-104) does not fold into a well-defined structure. Nevertheless, the "G/F" region has been found to have an effect on the packing of the helices in the "J-domain" in DnaJ(1-104). Particularly, the interhelical angles between Helix IV and other helices are significantly different in the two structures. In addition, there are some local conformational changes in the loop region connecting the two central helices. These structural differences in the "J-domain" in the presence of the "G/F" region may be related to the observation that DnaJ (1-78) is incapable of stimulating the ATPase activity of the molecular chaperone protein DnaK despite evidence that sites mediating the binding of DnaJ to DnaK are located in the 1-78 segment.
Organizational Affiliation:
Department of Chemistry, Yale University, New Haven, Connecticut 06520-8107, USA.