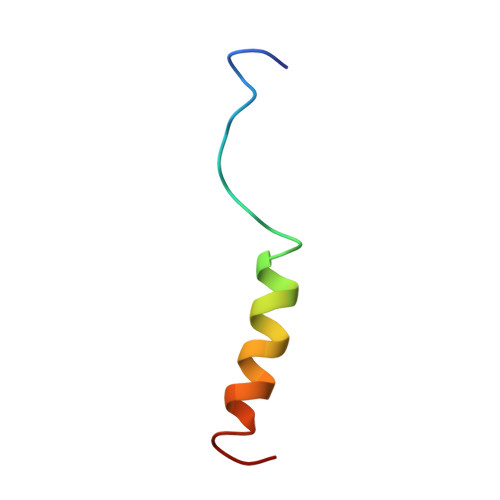
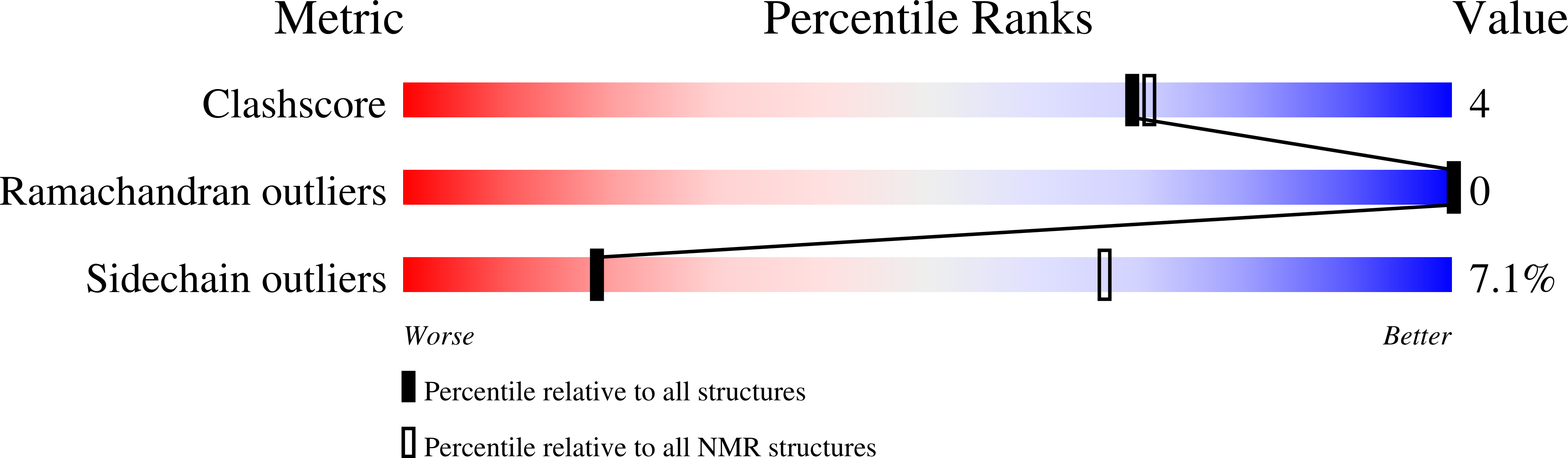Solution structures of human immunodeficiency virus type 1 (HIV-1) and moloney murine leukemia virus (MoMLV) capsid protein major-homology-region peptide analogs by NMR spectroscopy.
Clish, C.B., Peyton, D.H., Barklis, E.(1998) Eur J Biochem 257: 69-77
- PubMed: 9799104
- DOI: https://doi.org/10.1046/j.1432-1327.1998.2570069.x
- Primary Citation of Related Structures:
1BM4, 1BMX - PubMed Abstract:
The capsid domain of retroviral Gag proteins possesses a single highly conserved subdomain termed the major homology region (MHR). While the mutagenesis of residues in the MHR will impair virus infectivity, the precise solution structure and function of the MHR is not known. To aid the structure/function characterization of the MHR, the structures of synthetic peptides encompassing the MHR of the human immunodeficiency virus type I (HIV-1) and Moloney murine leukemia virus (MoMLV) capsid proteins were investigated by several techniques. Homology-based secondary-structure prediction suggested that the HIV-1 and MoMLV peptides could form 50% and 38% alpha-helix, respectively. CD studies indicated that, in the presence of 50% trifluoroethanol, the HIV-1 peptide adopts an alpha-helical structure over half of its length, while the MoMLV peptide is over one third alpha-helix. Further analysis by 1H-NMR suggested that the C-terminal portion of the MHR of each virus forms a helix in aqueous solution. Distance-geometry structures of each peptide were calculated from NOE distance restraints and were refined by restrained molecular dynamics. The C-terminal halves of both peptides were observed to be in an alpha-helical conformation, while the N-terminal halves were disordered. Furthermore, both helices were amphipathic with high conservation of amino acid side-chain character, suggesting that a conserved helical MHR C-terminus is essential to retroviral capsid protein function.
Organizational Affiliation:
Department of Chemistry, Portland State University, Oregon 97207-0751, USA.