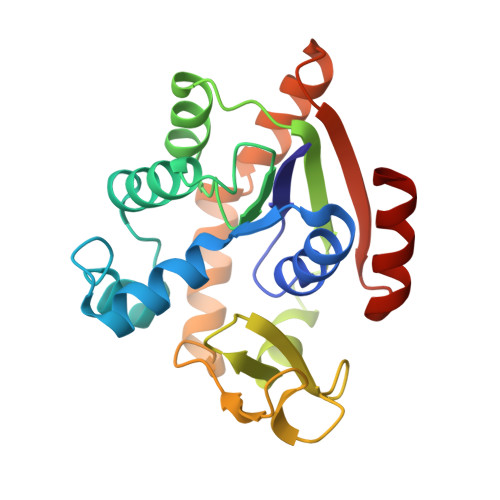
Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state.
Muller, C.W., Schulz, G.E.(1992) J Mol Biol 224: 159-177
- PubMed: 1548697
- DOI: https://doi.org/10.1016/0022-2836(92)90582-5
- Primary Citation of Related Structures:
1AKE - PubMed Abstract:
The structure of adenylate kinase from Escherichia coli ligated with the two-substrate-mimicking inhibitor P1,P5-bis(adenosine-5'-)pentaphosphate has been determined by X-ray diffraction and refined to a resolution of 1.9 A. The asymmetric unit of the crystals contains two copies of the complex, the structures of which agree well with each other. One of these copies is less well ordered in the crystals than the other, it shows generally higher temperature factors. The molecular packing in the crystals is discussed and correlated to crystal habit and anisotropic X-ray diffraction. The bound inhibitor simulates well the binding of substrates ATP and AMP, which are clearly assigned. The alpha-phosphate of AMP is well positioned for a nucleophilic attack on the gamma-phosphate of ATP. The observed structure readily allows the construction of a stabilized pentaco-ordinated transition state, as proposed for the known in-line mechanism of the enzyme, with nucleophile and leaving group in the apical positions of a trigonal bipyramid. The kinetic data of numerous mutations reported in the literature are correlated with the detailed structure of the enzyme. The mutants were classified. The concomitant increase of the Michaelis constants for ATP and AMP in the group of mutants that modify only the ATP-binding site cannot be explained.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie der Universität, Freiburg, Germany.