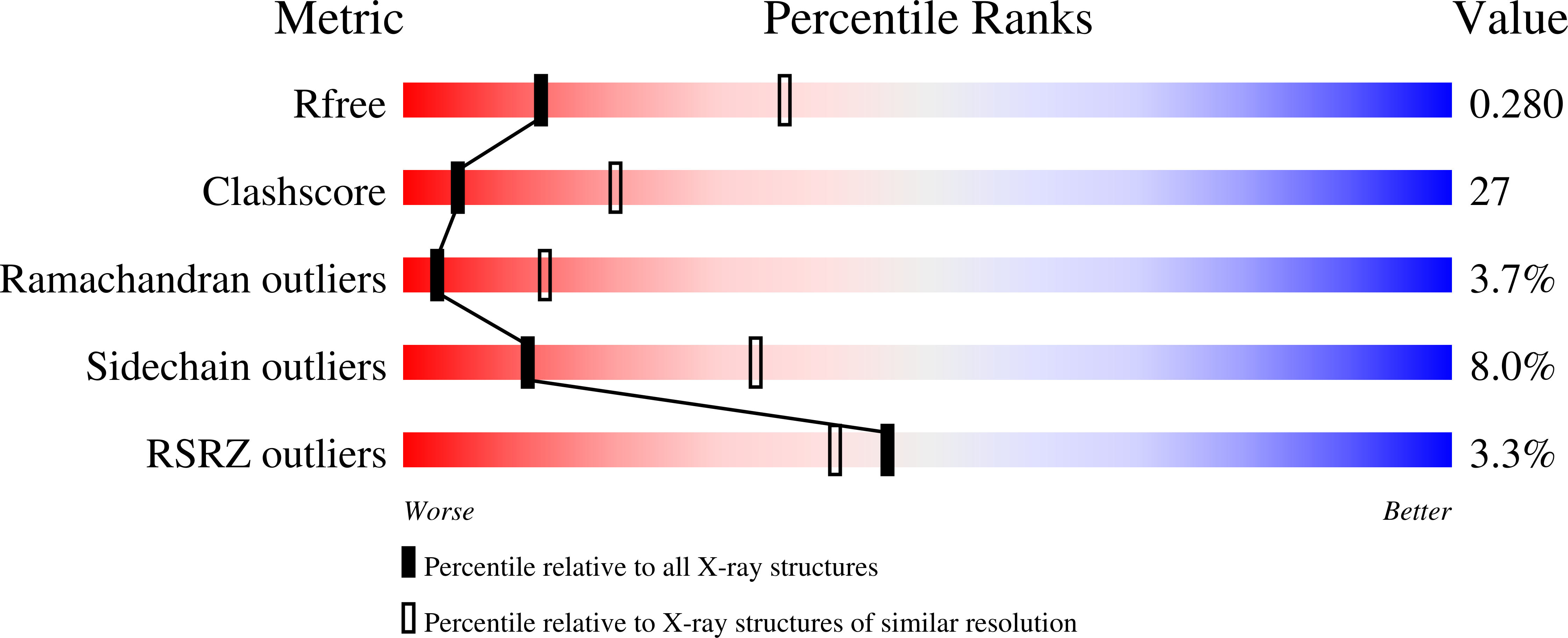
Conformational change of pseudouridine 55 synthase upon its association with RNA substrate
Phannachet, K., Huang, R.H.(2004) Nucleic Acids Res 32: 1422-1429
- PubMed: 14990747
- DOI: https://doi.org/10.1093/nar/gkh287
- Primary Citation of Related Structures:
1ZE1, 1ZE2 - PubMed Abstract:
Pseudouridine 55 synthase (Psi55S) catalyzes isomerization of uridine (U) to pseudouridine (Psi) at position 55 in transfer RNA. The crystal structures of Thermotoga maritima Psi55S, and its complex with RNA, have been determined at 2.9 and 3.0 A resolutions, respectively. Structural comparisons with other families of pseudouridine synthases (PsiS) indicate that Psi55S may acquire its ability to recognize a stem-loop RNA substrate by two insertions of polypeptides into the PsiS core. The structure of apo-Psi55S reveals that these two insertions interact with each other. However, association with RNA substrate induces substantial conformational change in one of the insertions, resulting in disruption of interaction between insertions and association of both insertions with the RNA substrate. Specific interactions between two insertions, as well as between the insertions and the RNA substrate, account for the molecular basis of the conformational change.
Organizational Affiliation:
Department of Biochemistry, School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, IL 61801, USA.