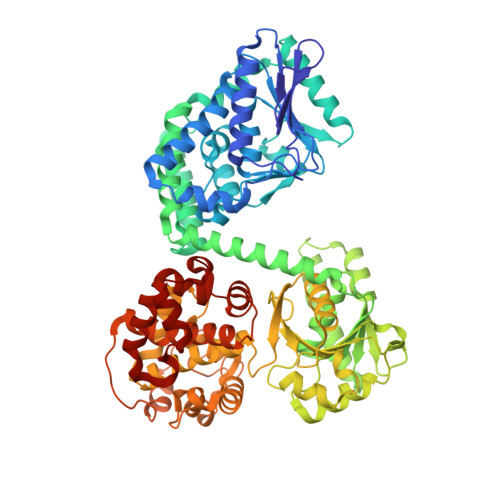
Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex
Ishikawa, M., Tsuchiya, D., Oyama, T., Tsunaka, Y., Morikawa, K.(2004) EMBO J 23: 2745-2754
- PubMed: 15229654
- DOI: https://doi.org/10.1038/sj.emboj.7600298
- Primary Citation of Related Structures:
1WDK, 1WDL, 1WDM - PubMed Abstract:
The atomic view of the active site coupling termed channelling is a major subject in molecular biology. We have determined two distinct crystal structures of the bacterial multienzyme complex that catalyzes the last three sequential reactions in the fatty acid beta-oxidation cycle. The alpha2beta2 heterotetrameric structure shows the uneven ring architecture, where all the catalytic centers of 2-enoyl-CoA hydratase (ECH), L-3-hydroxyacyl-CoA dehydrogenase (HACD) and 3-ketoacyl-CoA thiolase (KACT) face a large inner solvent region. The substrate, anchored through the 3'-phosphate ADP moiety, allows the fatty acid tail to pivot from the ECH to HACD active sites, and finally to the KACT active site. Coupling with striking domain rearrangements, the incorporation of the tail into the KACT cavity and the relocation of 3'-phosphate ADP bring the reactive C2-C3 bond to the correct position for cleavage. The alpha-helical linker specific for the multienzyme contributes to the pivoting center formation and the substrate transfer through its deformation. This channelling mechanism could be applied to other beta-oxidation multienzymes, as revealed from the homology model of the human mitochondrial trifunctional enzyme complex.
Organizational Affiliation:
Biomolecular Engineering Research Institute, Furuedai, Suita, Osaka, Japan.