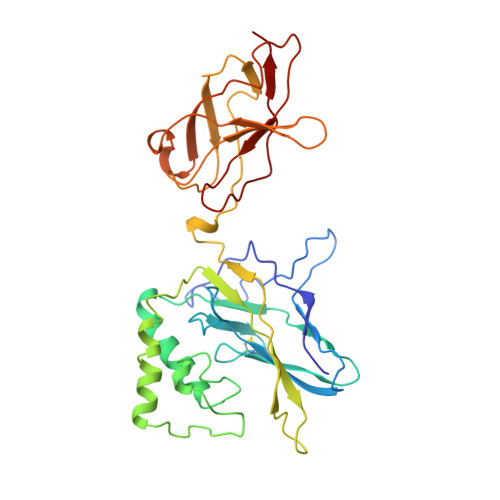
Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Chen, F.E., Huang, D.B., Chen, Y.Q., Ghosh, G.(1998) Nature 391: 410-413
- PubMed: 9450761
- DOI: https://doi.org/10.1038/34956
- Primary Citation of Related Structures:
1VKX - PubMed Abstract:
The NF-kappaB p50/p65 heterodimer is the classical member of the Rel family of transcription factors which regulate diverse cellular functions such as immune response, cell growth, and development. Other mammalian Rel family members, including the proteins p52, proto-oncoprotein c-Rel, and RelB, all have amino-terminal Rel-homology regions (RHRs). The RHR is responsible for the dimerization, DNA binding and cytosolic localization of these proteins by virtue of complex formation with inhibitor kappaB proteins. Signal-induced removal of kappaB inhibitors allows translocation of dimers to the cell nucleus and transcriptional regulation of kappaB DNA-containing genes. NF-kappaB specifically recognizes kappaB DNA elements with a consensus sequence of 5'-GGGRNYYYCC-3' (R is an unspecified purine; Y is an unspecified pyrimidine; and N is any nucleotide). Here we report the crystal structure at 2.9 A resolution of the p50/p65 heterodimer bound to the kappaB DNA of the intronic enhancer of the immunoglobulin light-chain gene. Our structure reveals a 5-base-pair 5' subsite for p50, and a 4-base-pair 3' subsite for p65. This structure indicates why the p50/p65 heterodimer interface is stronger than that of either homodimer. A comparison of this structure with those of other Rel dimers reveals that both subunits adopt variable conformations in a DNA-sequence-dependent manner. Our results explain the different behaviour of the p50/p65 heterodimer with heterologous promoters.
Organizational Affiliation:
Department of Biology, University of California, San Diego, La Jolla 92093-0359, USA.