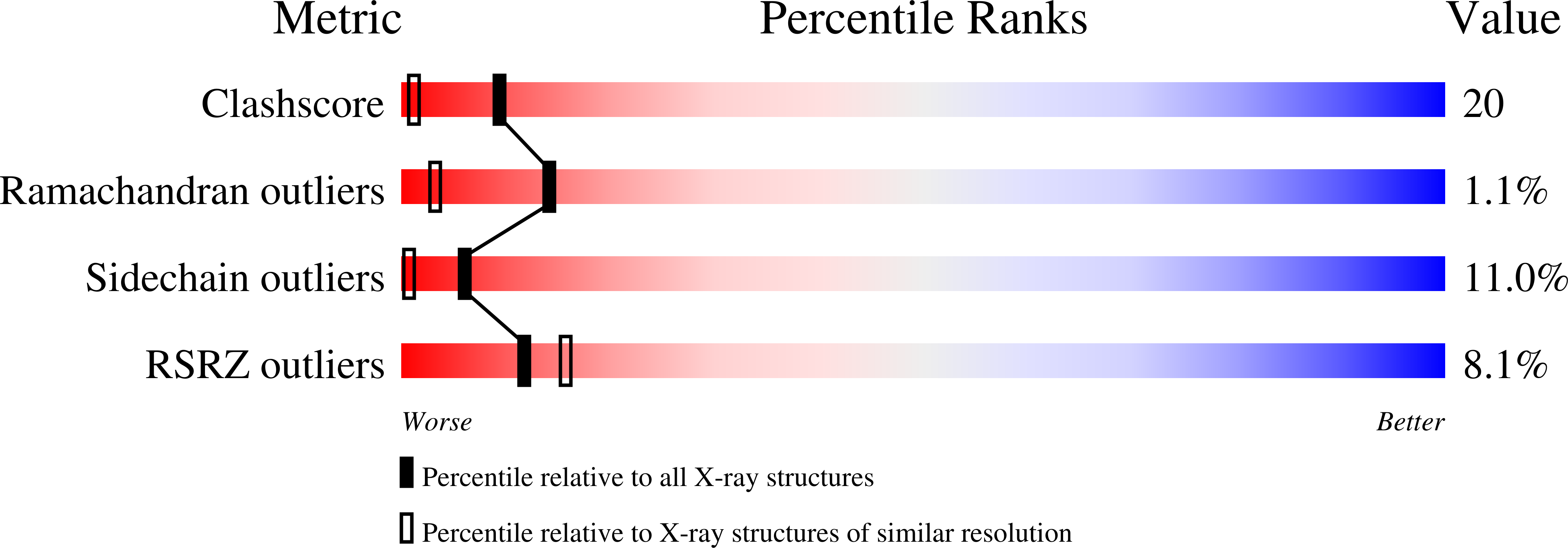The amino-terminal residues in the crystal structure of connective tissue activating peptide-III (des10) block the ELR chemotactic sequence.
Malkowski, M.G., Lazar, J.B., Johnson, P.H., Edwards, B.F.(1997) J Mol Biol 266: 367-380
- PubMed: 9047370
- DOI: https://doi.org/10.1006/jmbi.1996.0796
- Primary Citation of Related Structures:
1TVX - PubMed Abstract:
alpha-Chemokines comprise a family of cytokines that are chemotactic for neutrophils and have a structure similar to platelet factor 4 (PF4), in which the first two cysteine residues are separated by one residue (Cys-X-Cys). The two alpha-chemokines, connective tissue activating peptide-III (CTAP-III) and neutrophil activating peptide-2 (NAP-2), are carboxyl-terminal fragments of platelet basic protein (PBP) that are generated by monocyte-derived proteases. NAP-2 strongly stimulates neutrophils that are present during inflammation whereas its precursors, PBP and CTAP-III, are inactive, although they also possess the highly conserved, amino-terminal sequence, Glu-Leu-Arg (ELR), that is critical for receptor binding. To resolve this conundrum, we have determined the crystal structure of recombinant Asp-CTAP, which has ten fewer amino-terminal residues than CTAP-III but five more than NAP-2. The space group is P2(1)with unit cell dimensions a = 43.8 A, b = 76.8 A, c = 43.8 A, and beta =97.0 degrees, and a tetramer in the asymmetric unit. The molecular replacement method, with the NAP-2 tetramer as a starting model, was used to determine the initial phase information. The final R-factor is 0.196 (Rfree = 0.251) for 2sigma data from 7.0 to 1.75 A resolution. This high-resolution model of Asp-CTAP is the longest defined structure of an alpha-chemokine to date. The electron density map shows an over-all structure for Asp-CTAP that is very similar to that of NAP-2, but with the additional five amino-terminal residues folding back through a type-II turn, thereby stabilizing the oligomeric "inactive" state, and masking the critical ELR receptor binding region that is exposed in the structure of NAP-2.
Organizational Affiliation:
Department of Biochemistry, Wayne State University, Canfield, Detroit, MI 48201, USA.