Concanavalin A distorts the beta-GlcNAc-(1-->2)-Man linkage of beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man upon binding.
Moothoo, D.N., Naismith, J.H.(1998) Glycobiology 8: 173-181
- PubMed: 9451027
- DOI: https://doi.org/10.1093/glycob/8.2.173
- Primary Citation of Related Structures:
1TEI - PubMed Abstract:
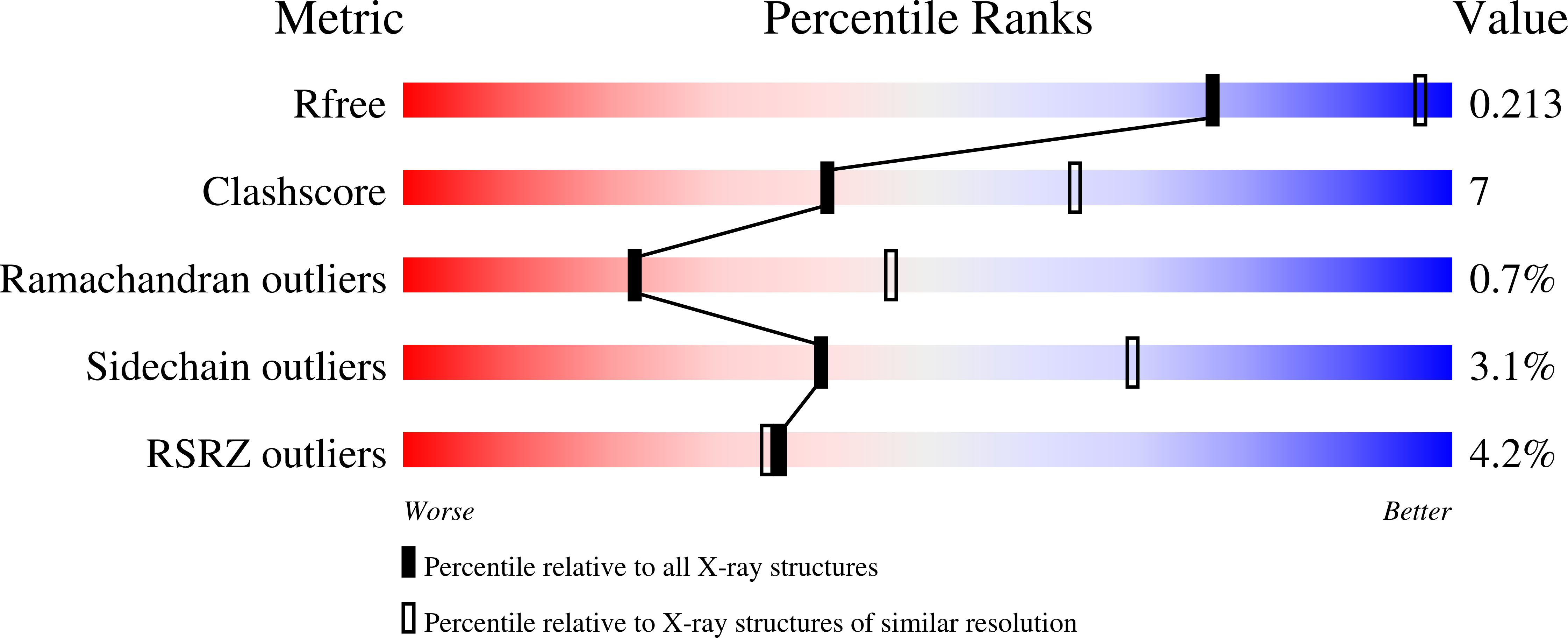
Carbohydrate recognition by proteins is a key event in many biological processes. Concanavalin A is known to specifically recognize the pentasaccharide core (beta-GlcNAc-(1-->2)-alpha- Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man-(1-->6)]-Man) of N-linked oligosaccharides with a Ka of 1.41 x 10(6 )M-1. We have determined the structure of concanavalin A bound to beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man to 2.7A. In six of eight subunits there is clear density for all five sugar residues and a well ordered binding site. The pentasaccharide adopts the same conformation in all eight subunits. The binding site is a continuous extended cleft on the surface of the protein. Van der Waals interactions and hydrogen bonds anchor the carbohydrate to the protein. Both GlcNAc residues contact the protein. The GlcNAc on the 1-->6 arm of the pentasaccharide makes particularly extensive contacts and including two hydrogen bonds. The binding site of the 1-->3 arm GlcNAc is much less extensive. Oligosaccharide recognition by Con A occurs through specific protein carbohydrate interactions and does not require recruitment of adventitious water molecules. The beta-GlcNAc-(1-->2)-Man glycosidic linkage PSI torsion angle on the 1-->6 arm is rotated by over 50 degrees from that observed in solution. This rotation is coupled to disruption of interactions at the monosaccharide site. We suggest destabilization of the monosaccharide site and the conformational strain reduces the free energy liberated by additional interactions at the 1-->6 arm GlcNAc site.
Organizational Affiliation:
Centre for Biomolecular Sciences, The University, St. Andrews, Scotland, United Kingdom.