Structural insights and functional implications of choline acetyltransferase
Govindasamy, L., Pedersen, B., Lian, W., Kukar, T., Gu, Y., Jin, S., Agbandje-McKenna, M., Wu, D.(2004) J Struct Biol 148: 226-235
- PubMed: 15477102
- DOI: https://doi.org/10.1016/j.jsb.2004.06.005
- Primary Citation of Related Structures:
1T1U - PubMed Abstract:
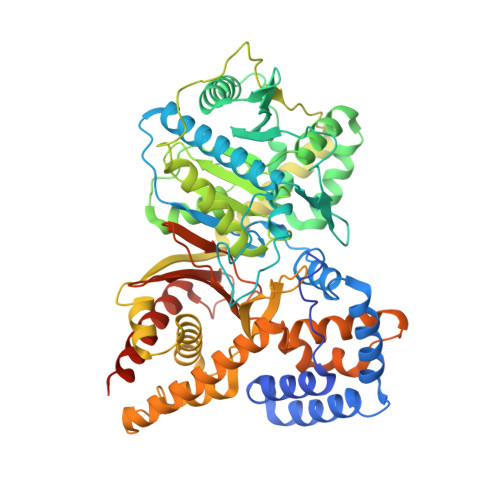
The biosynthetic enzyme for the neurotransmitter acetylcholine, choline acetyltransferase (ChAT) (E.C. 2.3.1.6), is essential for the development and neuronal activities of cholinergic systems involved in many fundamental brain functions. ChAT catalyzes the transfer of an acetyl group from acetyl-coenzyme A to choline to form the neurotransmitter acetylcholine. Since its discovery more than 60 years ago much research has been devoted to the kinetic studies of this enzyme. For the first time we report the crystal structure of rat ChAT (rChAT) to 1.55 A resolution. The structure of rChAT is a monomer and consists of two domains with an interfacial active site tunnel. This structure, with the modeled substrate binding, provides critical insights into the molecular basis for the production of acetylcholine and may further our understanding of disease causing mutations.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, McKnight Brain Institute and University of Florida, Gainesville, FL 32610, USA.