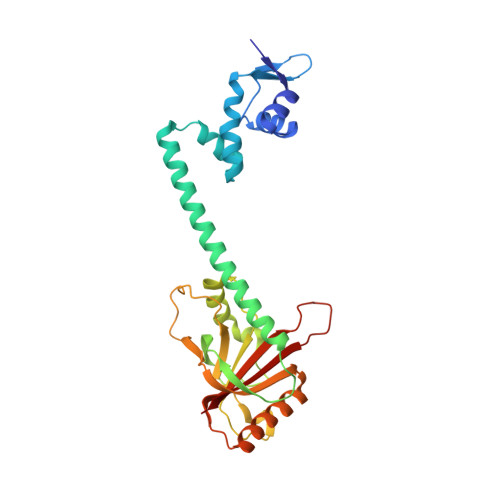
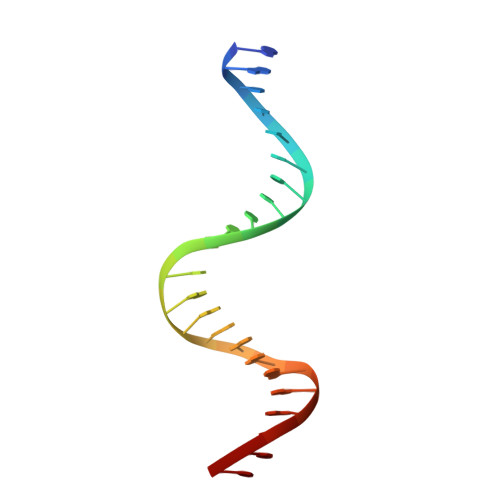
The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
Newberry, K.J., Brennan, R.G.(2004) J Biol Chem 279: 20356-20362
- PubMed: 14985361
- DOI: https://doi.org/10.1074/jbc.M400960200
- Primary Citation of Related Structures:
1R8D, 1R8E - PubMed Abstract:
Transcription regulators of the MerR family respond to myriad stress signals to activate sigma70/sigmaA-targeted genes, which contain suboptimal 19-bp spacers between their -35 and -10 promoter elements. The crystal structure of a BmrR-TPP(+)-DNA complex provided initial insight into the transcription activation mechanism of the MerR family, which involves base pair distortion, DNA undertwisting and shortening of the spacer, and realignment of the -35 and -10 boxes. Here, we describe the crystal structure of MerR family member MtaN bound to the mta promoter. Although the global DNA binding modes of MtaN and BmrR differ somewhat, homologous protein-DNA interactions are maintained. Moreover, despite their different sequences, the mta promoter conformation is essentially identical to that of the BmrR-TPP(+)-bound bmr promoter, indicating that this DNA distortion mechanism is common to the entire MerR family. Interestingly, DNA binding experiments reveal that the identity of the two central bases of the mta and bmr promoters, which are conserved as either a thymidine or an adenine in nearly all MerR promoters, is not important for DNA affinity. Comparison of the free and DNA-bound MtaN structures reveals that a conformational hinge, centered at residues N-terminal to the ubiquitous coiled coil, is key for mta promoter binding. Analysis of the structures of BmrR, CueR, and ZntR indicates that this hinge may be common to all MerR family members.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Oregon Health & Science University, Portland, OR 97239-3098, USA.