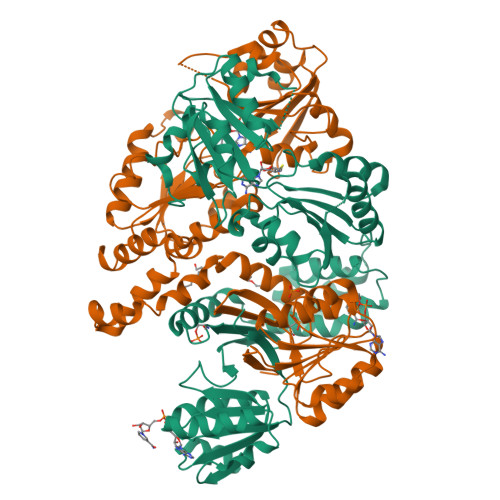
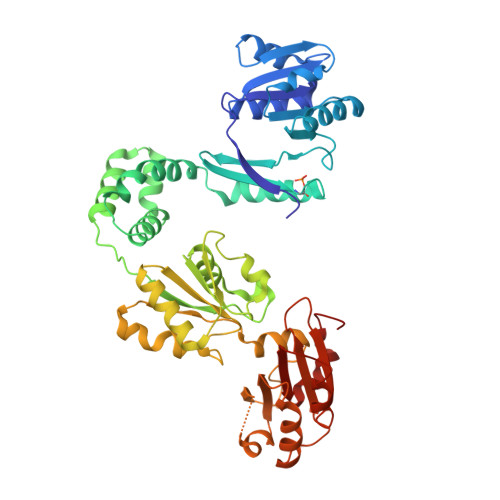
CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
Stroupe, M.E., Leech, H.K., Daniels, D.S., Warren, M.J., Getzoff, E.D.(2003) Nat Struct Biol 10: 1064-1073
- PubMed: 14595395
- DOI: https://doi.org/10.1038/nsb1007
- Primary Citation of Related Structures:
1PJQ, 1PJS, 1PJT - PubMed Abstract:
Sulfur metabolism depends on the iron-containing porphinoid siroheme. In Salmonella enterica, the S-adenosyl-L-methionine (SAM)-dependent bismethyltransferase, dehydrogenase and ferrochelatase, CysG, synthesizes siroheme from uroporphyrinogen III (uro'gen III). The reactions mediated by CysG encompass two branchpoint intermediates in tetrapyrrole biosynthesis, diverting flux first from protoporphyrin IX biosynthesis and then from cobalamin (vitamin B(12)) biosynthesis. We determined the first structure of this multifunctional siroheme synthase by X-ray crystallography. CysG is a homodimeric gene fusion product containing two structurally independent modules: a bismethyltransferase and a dual-function dehydrogenase-chelatase. The methyltransferase active site is a deep groove with a hydrophobic patch surrounded by hydrogen bond donors. This asymmetric arrangement of amino acids may be important in directing substrate binding. Notably, our structure shows that CysG is a phosphoprotein. From mutational analysis of the post-translationally modified serine, we suggest a conserved role for phosphorylation in inhibiting dehydrogenase activity and modulating metabolic flux between siroheme and cobalamin pathways.
Organizational Affiliation:
Department of Molecular Biology and Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, California 92037, USA.