Crystal structure of C-phycocyanin from Cyanidium caldarium provides a new perspective on phycobilisome assembly.
Stec, B., Troxler, R.F., Teeter, M.M.(1999) Biophys J 76: 2912-2921
- PubMed: 10354419
- DOI: https://doi.org/10.1016/S0006-3495(99)77446-1
- Primary Citation of Related Structures:
1PHN - PubMed Abstract:
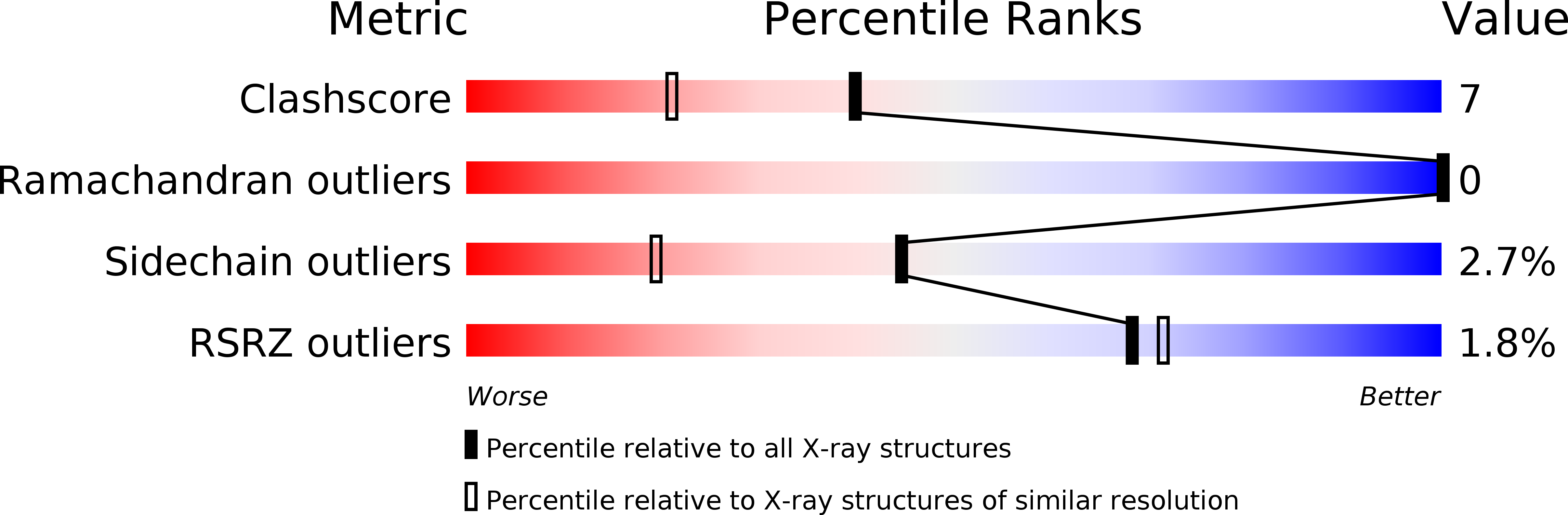
The crystal structure of the light-harvesting protein phycocyanin from the cyanobacterium Cyanidium caldarium with novel crystal packing has been solved at 1.65-A resolution. The structure has been refined to an R value of 18.3% with excellent backbone and side-chain stereochemical parameters. In crystals of phycocyanin used in this study, the hexamers are offset rather than aligned as in other phycocyanins that have been crystallized to date. Analysis of this crystal's unique packing leads to a proposal for phycobilisome assembly in vivo and for a more prominent role for chromophore beta-155. This new role assigned to chromophore beta-155 in phycocyanin sheds light on the numerical relationships among and function of external chromophores found in phycoerythrins and phycoerythrocyanins.
Organizational Affiliation:
Department of Chemistry, Merkert Chemistry Building, Boston College, Chestnut Hill, Massachusetts 02167, USA. stec@bioc.rice.edu